Comparative analysis of the gut microbiome’s profile of mothers and newborns with preterm and timely birth
Objective: To study the gut microbiota profile in women with preterm and term infants using next-generation sequencing (NGS).Gorina K.A., Khodzhaeva Z.S., Shubina Je., Rogacheva M.S., Muminova К.Т., Kutsar E.I., Priputnevich T.V.
Materials and methods: A prospective cohort study of 30 mother-infant pairs was conducted. The women and newborns were divided into two groups depending on gestational age (GA) at birth; group I consisted of 15 mother-infant pairs with spontaneous preterm birth (PTB), the median GA was 32.8 (2.08) weeks, and group II included 15 mother-infant pairs with term delivery (TD) at 39.5 (0.98) weeks of gestation. Fecal samples were analyzed using NGS.
Results: Meconium of newborns in both groups was not sterile; the dominant microorganisms were Acinetobacter and Cutibacterium (Propionibacteriaceae). Members of Bifidobacterium (Actinobacteria) were the most common microbes in maternal gut microbiota. Comparative analysis of the gut microbiota profile in mothers and infants in both groups did not show statistically significant differences (p>0.05).
Conclusion: Meconium was not sterile; it was antenatally colonized through maternal-fetal transmission, and a unique gut microbiota was formed. It should be interpreted as a new important factor of perinatal programming.
Keywords
Мaternal microbiome is as a key determinant of a number of important indicators of maternal and child health and, together with other perinatal factors, represents a basic aspect of perinatal programming [1].
The gut microbiota plays a pluripotent and not fully understood role in health and in various diseases, including immunomodulatory function (GALT – gut-associated lymphoid tissue) with colonization resistance [2]. The high diversity of species of microorganisms of the gut microbiota (GM) is associated with high resistance against pathogenic flora. A Norwegian study stated that women with preterm labor had lower content of Bifidobacterium and Streptococcus. Analysis of maternal GM using a metagenomic approach also showed a connection between poor GM and spontaneous preterm birth (PTB) [3]. The species diversity and the total amount of bacteria in meconium increases with gestational age, which may be associated with an increase in the duration of maternal-fetal metabolic communication [4].
According to modern concepts, gut colonization of the newborn begins in the antenatal period and is crucial in the development of the immune system and metabolism in general [5]. The previously accepted hypothesis that meconium is sterile and can be potentially colonized only by intra- and postnatal way is currently debatable, same as the “paradigm of a sterile uterus” [6, 7].
Most of the microorganisms from the huge species diversity of GM are represented by nonculturable forms of bacteria that cannot be “grown up” in growth medium, isolated in pure culture, and studied for properties [8]. To systematize nonculturable bacteria, the term “phylotype” is used, which characterizes microorganisms that are determined only by a sequence of the small subunit of 16S ribosomal RNA (16S rRNA) [9]. In metagenomic studies of microbial communities present in a sample (substrate), the most widely used approach is based on comparing the nucleotide sequences of the gene responsible for 16S rRNA synthesis [10]. However, in the last 2–3 years, researchers have started to use the complete sequence of the 16S rRNA gene to determine the composition of microbial communities. This is primarily due to the ability of NGS platforms (massively parallel/high throughput sequencing) to analyze long-reads, and skip the cloning step; amplified fragments from dozens of samples of biological material are sequenced simultaneously [11, 12]. The sequence of the 16S rRNA gene and its hypervariable regions has already been determined in a significant number of bacteria and is available in the Greengenes, Ribosomal Database Project (RDP), and SILVA databases [12–14].
The aim of the study was to describe the GM profile of mother and infants using the next-generation sequencing (NGS) in preterm and term birth.
Materials and methods
In a prospective study, 30 mother-infant pairs were examined. Women and newborns were divided into two groups depending on the gestational age at birth. Group I included 15 mother-infant pairs with spontaneous preterm birth at 32.8 (2.08) weeks; group II included of 15 mother-infant pairs with term delivery (TD) at 39.5 (0.98) weeks.
All patients included in the study were analyzed for anamnestic, clinical and laboratory data: somatic and obstetric-gynecological history, assessment of pregnancy, a standard panel of laboratory parameters of peripheral blood, assessment of the microbial composition of the lower genital tract secrete; functional assessment and fetometry of the fetus. After labor, delivery methods and perinatal outcomes were analyzed. We assessed the Apgar scores of newborns, the course of early and late neonatal periods, and analyzed clinical, laboratory, and instrumental data.
The inclusion criteria for the control group and group of comparison were a singleton and spontaneous pregnancy; for group with PTB – gestational age 280–346 weeks, and signed informed consent to participate in the study, including application of special methods for scientific purposes to perform standard clinical tests in the infants. To minimize the influence of confounders, patients with severe obstetric (except for the presence of PTB, late pregnancy loss and habitual failure to conceive in the history) and extragenital pathology; carriers of Streptococcus agalactiae (Group B Streptococcus) confirmed by culture identification were not included in the study.
The exclusion criteria were: multiple pregnancies, placenta praevia/accreta, diagnosed chronic inflammatory bowel disease.
All patients signed an informed consent to participate in the study. The study was approved by the Ethics Committee of the Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation.
The GM species composition was analyzed by NGS using V3-V4 region. The selected region was amplified with primers 357F and 806R, DNA was extracted with PREP-RAPID DNA Extraction Kit v2, and 16S RNA gene sequence was amplified with the DTprime amplifier (“DNA-Technology LLC”, Russia). The quality of the amplicons was assessed in 2% agarose gel. The quality and concentration of DNA libraries for NGS sequencing were checked using a Bioanalyzer. Sequencing was performed using MiSeq (Illumina, USA) on v2 flow cell according to the manufacturer's instructions. Data analysis was performed using the QIMME software package.
Statistical analysis
Statistical analysis of the data was performed using GraphPad Prism 8.3 and IBM SPSS Statistics 22 software. Quantitative data with a normal distribution are presented as arithmetic mean and standard deviation; for distributions other than normal, the data are presented as median and quartiles. The D'Agostino–Pearson Omnibus Test, the Anderson–Darling test, the Kolmogorov–Smirnov test and Levene's test were used to determine normality of distribution. The following data processing methods were used: Fisher's exact test, comparative analysis of variables using parametric Student's t-test for independent samples; in the absence of normal distribution, the non-parametric statistics methods – the Mann–Whitney U test and the Wilcoxon signed-rank test – were used. Differences were considered statistically significant at p<0.05.
Results
The study included 30 mother-infant pairs. According to gestational age at the time of delivery, the pairs were divided into two groups (PTB and TD), comparable in age, body weight, height, and body mass index (BMI). The clinical characteristics of the patients are shown in the Table.
The analysis of the clinical and medical history parameters presented in the table demonstrated typical risk factors for PTB. Miscarriages and a history of PTB were significantly more common in patients with PTB. The rate of such factors as smoking, PCOS, endometriosis, uterine myoma, gestational diabetes mellitus, preeclampsia, and various extragenital pathologies did not differ significantly in both groups (p>0.05). Gastrointestinal diseases: chronic gastritis, duodenitis, gastric ulcers, biliary dyskinesia, and inflammatory bowel disease were found significantly more often in the group of patients with PTB. To evaluate the dietary habits and bowel function, we conducted a questionnaire survey of patients in both groups. No significant differences were found in the characteristics of the diet (5 options were offered): a diverse diet, including unhealthy foods (fast food, chips, etc.) (5/15 (33.3%) and 8/15 (53, 3%), respectively, p=0.46); diverse healthy diet (10/15 (66.7%) and 5/15 (33.3%), respectively, p=0.14); without carbohydrates (0/15 and 1/15 (6.7%), respectively, p>0.99); without proteins and fats (none of the respondents chose this variant). Changes in bowel motorics (constipation) were significantly more common in the PTB group (9/15 (60%) and 2/15 (13.3%), respectively; p=0.02, RR 2.59; 95% CI 1.29–5.52). Smokers (3/15 (20%) and 2/15 (13.3%), respectively; p>0.99), and pet owners (8/15 (53.3%) and 5/15 (33.3%), respectively; p=0.46) were evenly represented in both groups.
The analysis of the course of pregnancy in patients with PTB showed a statistically significantly higher rate of PTB threatening episodes in the III trimester of pregnancy (11/15 (73.3%) and 0/15 (0%), respectively; p<0.01, RR 2.67; 95% CI 2.13–4.44), and a higher rate of cervical insufficiency requiring treatment (cerclage and/or pessary; 9/15 (60%) and 1/15 (6.7%), respectively; p<0.01, RR 3.00; 95% CI 1.54–6.31). An analysis of antibacterial therapy in studied women showed a statistically significant difference between the groups in the II trimester (9/15 (60%) and 1/15 (6.7%), respectively; p<0.01, RR 3.00; 95% CI 1.54–6.31). The main indications for antibiotic therapy were respiratory disease and abundant growth of pathogens in the vagina. However, there were no significant differences in the I trimester (2/15 (13.3%) and 0/15 (0%), respectively; p>0.05) and III trimester (2/15 (13.3%) and 1/15 (6.7%), respectively; p>0.05) of pregnancy when analyzing this indicator.
In a comparative analysis of delivery patterns, regardless of gestational age, the predominant method in both groups was vaginal delivery (10/15 (66.7%) and 11/15 (73.3%), respectively; p>0.05). The rate of premature rupture of membranes was higher in the PTB group (5/15 (33.3%) and 1/15 (6.7%), respectively), but was not statistically significant (p>0.05).
In newborns, in addition to differences in characteristics dependent on gestational age, such as Apgar scores (7.0 [6; 8] and 8 [8; 8], p<0.01 for the 1st minute; 8.0 [7; 8] and 9 [9; 9], p<0.001, for the 5th minute) and anthropometric data (mean body weight 2078 g (575.0) and 3431 g (340.8), p<0.001), there was a higher and statistically significant prevalence of perinatal-specific infections (10/15 (66.7%) and 0/15 (0%), respectively; p<0.001, RR 4.00; 95% CI 3.59–6.85). The Silverman-Andersen Retraction score for respiratory distress in preterm infants was 2.0 [2; 3]. Also, preterm infants had a higher incidence of complications due to morphofunctional dismaturity: newborn respiratory distress syndrome – 10/15 (66.7%), transient tachypnea of the newborn – 3/15 (20%), congenital pneumonia – 6/15 (40%), retinopathy – 2/15 (13.3%).
Features of GM profile (the most represented types – more than 1%) of all patients included in the study (30 mother-infant pairs) are shown in Figure 1 as a graph with bar charts.
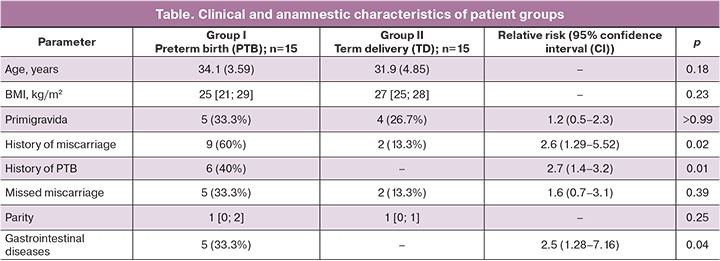
Studies on animals have shown the possibility of vertical intrauterine transmission of microbes to the fetus, and similar mechanisms can exist in humans [5]. In our study, the meconium was also not sterile; among newborns, the dominant microorganisms were represented by Propionibacterium genus or propionic acid bacteria, which are normal flora of the human gastrointestinal tract. In women, the dominant representatives of GM in both groups were microorganisms of the Bifidobacterium genus (type Actinobacteria) and the family Lachnospiraceae (spore-forming bacteria of the order Clostridiales), which are the most common taxa of gut microbiota.
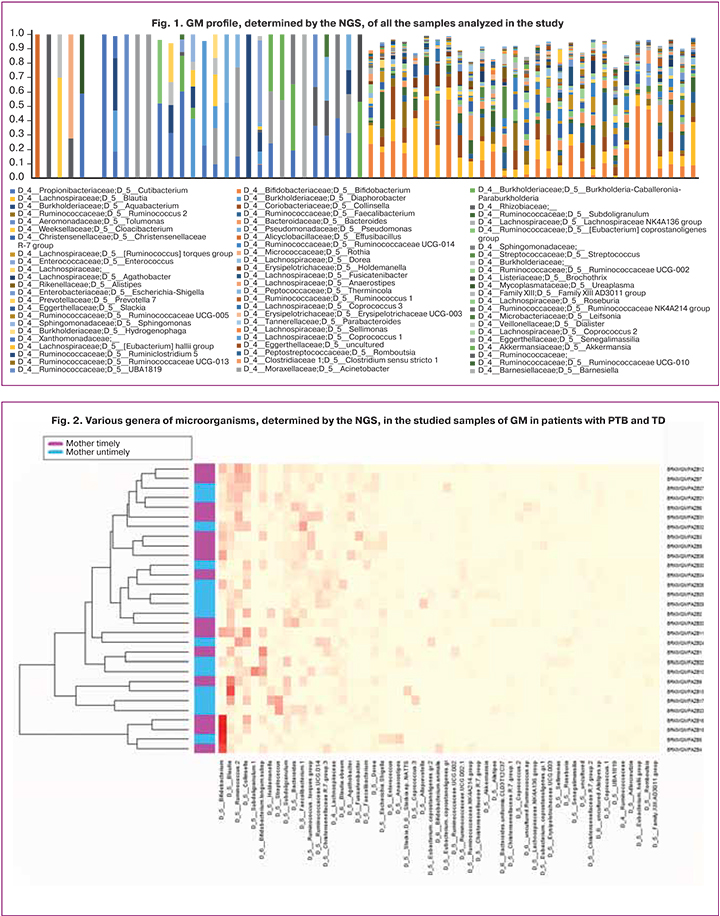
Based on the common hypothesis on the involvement of GM in the development of the great obstetrical syndromes and its impact on perinatal outcomes [15], the participants of the study were divided into two groups depending on the gestational age at the time of delivery. The data on GM sequencing of the pregnant women with PTB and TD are presented in Figure 2, for full-term and preterm infants – in Figure 3. The most common genus of bacteria (prevalence above 5.0%) were determined by sequencing of the 16S ribosomal RNA fragment.
According to the results, the major representatives of the GM in both groups were microorganisms of the genus Bifidobacterium (Actinobacteria type), gram-positive anaerobes responsible for the health of the human large intestine [16]. It is worth noting that with the same taxonomic diversity, patients with PTB had some “depletion” of almost all species of the normal microbiota.
Acinetobacter and Cutibacterium species (members of the Propionibacteriaceae family) were most abundant in the meconium of newborns. Acinetobacter is a saprophytic microorganism presented in healthy humans on the skin, in gut, and in the genitourinary system. For a long time, Acinetobacter spp. were considered low-pathogenic, but later it was proved that under certain conditions their virulence can significantly increase and lead to the development of severe infections, including meningitis and sepsis [17].
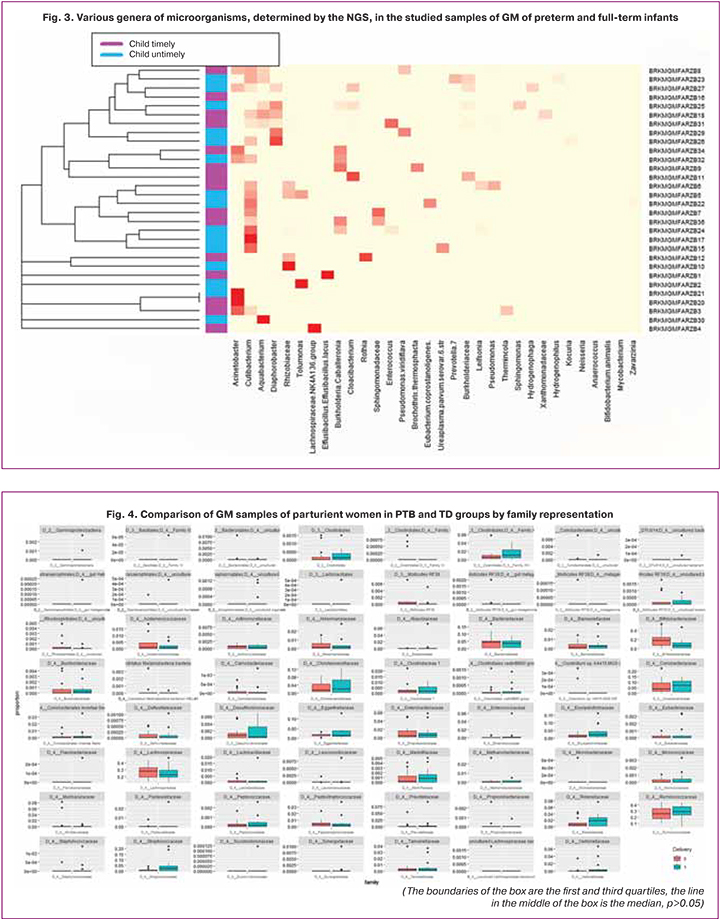
Despite numerous literature data confirming the association of dysbiosis with PTB [3, 18, 19], a statistical comparison of the medians of the groups with PTB and TD showed no significant differences. The absence of statistically significant differences when using the Wilcoxon signed-rank test is also observed when comparing groups by family representation in parturient women (p>0.05, median ratio 1.74–6.4; Fig. 4), which may be due to the small size of the studied groups.
Discussion
The human body contains trillions of cells, most of which are xenobiotic microorganisms. The Human Microbiome Project demonstrated the vastness of the microbial world, which has been underestimated for a long time, and confirmed many causal relationships between the microbiota of various organs and systems and physiological and pathological conditions [20]. The metagenomic approach, based on the use of high throughput sequencing, is currently considered to be the most accurate and detailed method of microbiota assessment, allowing the identification of previously unknown microorganisms with their potential clinical significance [11]. However, this method has a number of disadvantages and limitations, primarily, time and labor stage, along with high costs.
In our study, no statistically significant differences were found in GM profiles of mothers and newborns in PTB and TD; this does not confirm the absence of a pathogenetic relationship, which has been shown many times in foreign publications [3, 18], and had been proved in our previous study, but using the culture technique [19]. The lack of statistical significance is most likely due to the small size of the group, so it requires further research.
The most important finding of our study was the evidence of the non-sterility of meconium, which supports the theory of intrauterine microbial colonization of the fetus through maternal-fetal transmission [7, 21]. The exact mechanism of bacterial transmission from mother to fetus is unknown, but it is the gastrointestinal tract of the mother that is currently considered the most likely source [22]. Understanding the timing and mechanisms involved in the first colonization of the human gut is of fundamental importance for a comprehensive analysis of the impact of GM on newborn development [5, 21].
Conclusion
The gut microbiota plays a multifunctional role and is not limited to maintaining gastrointestinal homeostasis. During pregnancy, together with other factors, it influences formation of the microbiome of the newborn. The non-sterility of meconium, its antenatal colonization through maternal-fetal transmission with the formation of a unique microbiota, should be regarded as a new important factor in perinatal programming. Over the past two decades, sequencing has taken on leadership in human microbiome research. The high sensitivity and phylogenetic resolution of the NGS method, as well as the new data obtained in the experiments, require further validation on larger samples before being introduced into clinical practice.
References
- Dunlop A.L., Mulle J.G., Ferranti E.P., Edwards S., Dunn A.B., Corwin E.J. Maternal microbiome and pregnancy outcomes that impact infant health: a review. Adv. Neonatal Care. 2015; 15(6): 377-85. https://dx.doi.org/10.1097/ANC.0000000000000218.
- Воронцова З.А., Никитюк Д.Б., Кудаева Э.Ф. Кишечно-ассоциированная лимфоидная ткань как информационно-корректирующая система экстремальных состояний (краткий обзор литературы). Вестник новых медицинских технологий. 2016; 4: 289-94. [Vorontsova Z.A., Nikityuk D.B., Kudaeva E.F. Gut associated lymphoid tissue as information and corrective system of extreme conditions (brief literature report). Journal of New Medical Technologies. 2016; 4. (in Russian)]. https://dx.doi.org/10.12737/21854.
- Dahl C., Stanislawski M., Iszatt N., Mandal S. Gut microbiome of mothers delivering prematurely shows reduced diversity and lower relative abundance of Bifidobacterium and Streptococcus. Abdo Z, editor. PLoS One. 2017; 12(10): e0184336. https://dx.doi.org/10.1371/journal.pone.0184336.
- Gibson M.K., Crofts T.S., Dantas G. Antibiotics and the developing infant gut microbiota and resistome. Curr. Opin. Microbiol. 2015; 27: 51-6. https://dx.doi.org/10.1016/j.mib.2015.07.007.
- Walker R.W., Clemente J.C., Peter I., Loos R.J.F. The prenatal gut microbiome: are we colonized with bacteria in utero? Pediatr. Obes. 2017; 12(Suppl. 1): 3-17. https://dx.doi.org/10.1111/ijpo.12217.
- Koleva P.T., Kim J.-S., Scott J.A., Kozyrskyj A.L. Microbial programming of health and disease starts during fetal life. Birth Defects Res. C Embryo Today. 2015; 105(4): 265-77. https://dx.doi.org/10.1002/bdrc.21117.
- Funkhouser L.J., Bordenstein S.R. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 2013; 11(8): e1001631. https://dx.doi.org/10.1371/journal.pbio.1001631.
- Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012; 18(12): 1185-93. https://dx.doi.org/10.1111/1469-0691.12023.
- Moreira D., López-García P. Phylotype. In: Encyclopedia of astrobiology. Berlin, Heidelberg: Springer; 2011: 1254. https://dx.doi.org/10.1007/978-3-642-11274-4_1210.
- Johnson J.S., Spakowicz D.J., Hong B.-Y., Petersen L.M., Demkowicz P., Chen L. et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019; 10(1): 5029. https://dx.doi.org/10.1038/s41467-019-13036-1.
- Malla M.A., Dubey A., Kumar A., Yadav S., Hashem A., Abd Allah E.F. Exploring the human microbiome: the potential future role of next-generation sequencing in disease diagnosis and treatment. Front. Immunol. 2019; 9: 2868. https://dx.doi.org/10.3389/fimmu.2018.02868.
- Алексеева А.Е., Бруснигина Н.Ф. Возможности и перспективы применения методов массивного параллельного секвенирования в диагностике и эпидемиологическом надзоре за инфекционными заболеваниями. МедиАль. 2014; 12(2): 6-28. [Alekseeva A.E., Brusnigina N.F. Possibilities and prospects of using methods of massive parallel sequencing in the diagnosis and epidemiological surveillance of infectious diseases. Medial. 2014; 2(12): 6-28. (in Russian)].
- Cole J.R., Wang Q., Fish J.A., Chai B., McGarrell D.M., Sun Y. et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014; 42(Database issue): D633-42. https://dx.doi.org/10.1093/nar/gkt1244.
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013; 41(Database issue): D590-6. https://dx.doi.org/10.1093/nar/gks1219.
- Solt I. The human microbiome and the great obstetrical syndromes: a new frontier in maternal-fetal medicine. Best Pract. Res. Clin. Obstet. Gynaecol. 2015; 29(2): 165-75. https://dx.doi.org/10.1016/j.bpobgyn.2014.04.024.
- Batt C.A., Tortorello M.L., eds. Encyclopedia of food microbiology. 2nd ed. 2014: 216-22.
- Zarrilli R., Bagattini M., Esposito E.P., Triassi M. Acinetobacter infections in neonates. Curr. Infect. Dis. Rep. 2018; 20(12): 48. https://dx.doi.org/10.1007/s11908-018-0654-5.
- Hiltunen H., Collado M.C., Ollila H., Kolari T., Tölkkö S., Isolauri E. et al. Spontaneous preterm delivery is reflected in both early neonatal and maternal gut microbiota. Pediatr Res. 2021 Aug 4. https://dx.doi.org/10.1038/s41390-021-01663-8. Online ahead of print.
- Горина К.А., Ходжаева З.С., Муравьева В.В., Муминова К.Т., Донников А.Е., Припутневич Т.В. Роль микробиоты кишечника матери при спонтанных преждевременных родах. Акушерство и гинекология. 2020; 8: 64-71. [Gorina K.A., Khodzhaeva Z.S., Muravieva V.V., Muminova К.Т., Donnikov A.E., Priputnevich T.V. The role of maternal gut microbiota in spontaneous preterm birth. Obstetrics and Gynecology. 2020; 8: 64-71. (in Russian)]. https://dx.doi.org/10.18565/aig.2020.8.64-71.
- Integrative HMP (iHMP) Research Network Consortium. The integrative human microbiome project. Nature. 2019; 569(7758): 641-8. https://dx.doi.org/10.1038/s41586-019-1238-8.
- Romano-Keeler J., Weitkamp J.-H. Maternal influences on fetal microbial colonization and immune development. Pediatr. Res. 2015; 77(1-2): 189-95. https://dx.doi.org/10.1038/pr.2014.163.
- Hu J., Nomura Y., Bashir A., Fernandez-Hernandez H., Itzkowitz S., Pei Z. et al. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS One. 2013; 8(11): e78257. https://dx.doi.org/10.1371/journal.pone.0078257.
Received 01.09.2021
Accepted 09.11.2021
About the Authors
Ksenia A. Gorina, Junior Researcher, 1st Obstetric Department of Pregnancy Pathology, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, +7(926)649-77-32, k_gorina@oparina4.ru, https://orcid.org/0000-0001-6266-2067,117997, Russia, Moscow, Ac. Oparin str., 4.
Zulfiya S. Khodzhaeva, Dr. Med. Sci., Professor, Deputy Director for Research of the Institute of Obstetrics, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, +7(916)407-75-67, zkhodjaeva@mail.ru,
https://orcid.org/0000-0001-8159-3714, 117997, Russia, Moscow, Ac. Oparin str., 4.
Jekaterina Shubina, Head of the Laboratory of Genomic Data Analysis, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, +7(495)531-44-44, e_shubina@oparina4.ru, https://orcid.org/0000-0003-4383-7428,
117997, Russia, Moscow, Ac. Oparin str., 4.
Margarita S. Rogacheva, Junior Researcher, Laboratory of Genomic Data Analysis, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, +7(953)370-85-12, m_rogacheva@oparina4.ru, https://orcid.org/0000-0002-2495-2554,
117997, Russia, Moscow, Ac. Oparin str., 4.
Kamilla T. Muminova, Ph.D., Junior Researcher, 1st Obstetric Department of Pregnancy Pathology, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, +7(916)373-77-07, k_muminova@oparina4.ru, https://orcid.org/0000-0003-2708-4366,
117997, Russia, Moscow, Ac. Oparin str., 4.
Elena I. Kutsar, resident doctor, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, +7(916)766-98-99, kutsar.elena@mail.ru, https:// orcid.org/0000-0002-6974-7773, 117997, Russia, Moscow, Ac. Oparin str., 4.
Tatyana V. Priputnevich, Dr. Med. Sci., Head of the Departament of Microbiology, Clinical Pharmacology and Epidemiology, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, +7(903)264-12-57, priputl@gmail.com,
https://orcid.org/0000-0002-4126-9730, 117997, Russia, Moscow, Ac. Oparin str., 4.
Author’s contributions: Gorina K.A., Khodzhaeva Z.S., Shubina Je., Rogacheva M.S., Muminova К.Т., Kutsar E.I.,
Priputnevich T.V. – design of the study, data collection and analysis, publications review on the topic of the article, editing the article; Gorina K.A., Khodzhaeva Z.S. – manuscript writing; Rogacheva M.S., Gorina K.A. – statistical analysis
of the obtained data.
Conflicts of interest: The authors declare no conflicts of interest.
Funding: The study was funded by Russian Foundation for Basic Research (RFBR), project No. 19-315-90104.
Patient Consent for Publication: Mothers of newborns provided informed consent for the publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator.
For citation: Gorina K.A., Khodzhaeva Z.S., Shubina Je., Rogacheva M.S., Muminova К.Т., Kutsar E.I., Priputnevich T.V. Comparative analysis of the gut microbiota profile
in mothers and term and preterm infants.
Akusherstvo i Gynecologia/Obstetrics and Gynecology. 2021; 12: 88-95 (in Russian)
https://dx.doi.org/10.18565/aig.2021.12.88-95



