The role of L1 and L2 capsid proteins of human papillomavirus in the differential diagnosis of squamous intraepithelial lesions
Dobrovolskaya D.A., Bayramova G.R., Asaturova A.V., Badlaeva A.S., Tregubova A.V.
Objective: To evaluate and analyze the expression of L1 and L2 proteins of human papillomavirus (HPV) using the techniques of immunohistochemistry (IHC) and L1 immunocytochemistry (ICC) for differential diagnosis of cervical squamous intraepithelial lesions.
Materials and methods: The cross-sectional study included 128 patients who presented to the Department of Outpatient Clinical Research Development of the National Medical Research Centre for Obstetrics, Gynecology and Perinatology in Moscow in the period from January 2022 to January 2023. The patients met the inclusion and exclusion criteria. Depending on the results of cervical cytology, all examined patients were divided into four groups: group 1 included 36 women with NILM cytology, group 2 consisted of 31 patients with ASC-US cytology, group 3 included 31 patients with LSIL cytology, and group 4 consisted of 30 patients with HSIL cytology. All patients underwent cytological, colposcopic and histological examination as well as IHC and ICC to determine the expression level of HPV L1 and L1/L2 viral proteins, respectively.
Results: The expression of HPV L1 was statistically significantly more frequent and revealed by ICC technique in the LSIL group (65.6%) compared to HSIL (20%) and NILM (19.4%) (Pearson’s χ2, p<0.001). LSIL was 2.1 times more likely to be detected by cytological examination among all L1-positive samples compared to L1-negative samples (95% CI: 1.98-10.87, p<0.001). The use of IHC technique made it possible to detect the expression of L1 viral protein in 72.9% of LSIL (CIN1) cases, which was statistically significantly more frequent than in other study groups (p<0.001). In the group of histologically verified LSIL (CIN1), positive expression of HPV L2 protein was observed in 48.8% of cases.
Conclusion: The use of HPV L1 and L2 capsid proteins can significantly improve the differential diagnosis of mild squamous intraepithelial lesions.
Authors’ contributions: Dobrovolskaya D.A., Bayramova G.R., Asaturova A.V. – developing the design of the study, review of publications on the issue of the study and their translation, statistical analysis of the obtained data, writing the text of the manuscript; Dobrovolskaya D.A., Badlaeva A.S., Tregubova A.V. – obtaining the data for analysis; Asaturova A.V., Bayramova G.R. – structuring and editing the article.
Conflicts of interest: The authors declare that there are no conflicts of interest.
Funding: The study was carried out with the financial support of the Russian Foundation for Basic Research and the Moscow City Government within the framework of scientific project No. 21-315-70048, “Study of the role of viral replicative early response proteins of human papillomavirus in modulation of signaling pathways of cervical epitheliocytes in initial forms of cervical intraepithelial squamous lesions using neural network analysis methods”.
Ethical Approval: The study was approved by the Ethical Review Board of the Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Moscow.
Patient Consent for Publication: The patients signed informed consent for the publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator.
For citation: Dobrovolskaya D.A., Bayramova G.R., Asaturova A.V., Badlaeva A.S., Tregubova A.V. The role of L1 and L2 capsid proteins of human papillomavirus in the differential diagnosis of squamous intraepithelial lesions.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2025; (2): 81-90 (in Russian)
https://dx.doi.org/10.18565/aig.2024.298
Keywords
Highly oncogenic types of human papillomavirus (HPV) have been shown to be major causative factors in the development of squamous intraepithelial lesions (SIL) and cervical cancer. It is known that HPV alone is not sufficient to induce malignant transformation of mild squamous cell intraepithelial lesions, and the assessment of SIL progression associated with different HPV types is controversial. Today, it is extremely important to search for biomarkers that will help to distinguish the ‘pool’ of patients with cytologically confirmed low-grade squamous intraepithelial lesion (LSIL) caused by transient HPV infection (which can be self-eliminated by the virus) from lesions associated with integration of viral DNA into the genome of host cells with a high potential for progression [1, 2]. According to the literature, the diagnostic accuracy of cytological examination for LSIL is 79.4%, while for high-grade squamous intraepithelial lesions (HSIL) it is 100% [3, 4]. In this context, new approaches have been developed to optimize the differential diagnosis of SIL in terms of HPV biology for the purpose of improving the quality of care [5, 6].
The aim of the study is to evaluate and compare the expression of HPV L1/L2 proteins using the techniques of immunohistochemistry (IHC) and L1 immunocytochemistry (ICC) for differential diagnosis of SIL.
Materials and methods
The cross-sectional study included 128 patients who presented to the Department of Outpatient Clinical Research Development of the National Medical Research Centre for Obstetrics, Gynecology and Perinatology in Moscow in the period from January 2021 to December 2022. All the patients were divided into four groups: group 1 (control group) included women with negative for intraepithelial lesion or malignancy (NILM) cytology (n=36), group 2 consisted of patients with atypical squamous cells of undetermined significance (ASC-US) cytology (n=31), group 3 included patients with LSIL cytology (n=31), and group 4 consisted of patients with HSIL cytology (n=30). HPV positive women aged between 18 and 45 years were eligible for including in the study. Non-inclusion criteria were pregnancy, lactation period, presence of neuropsychiatric diseases (inability to follow the protocol) and aggravation of oncological diseases. All patients included in the study signed informed consent and underwent cytological (liquid cytology), colposcopic and histological examinations, as well as ICH and ICC examinations to determine HPV L1 and L1/L2 protein expression levels, respectively.
To perform immunohistochemical study, 4-5 µm thick sections were prepared from paraffin blocks, which were then placed on SuperFrost Plus (Thermo Scientific) slides with a positive charge and dried in a thermostat for 24 h at 48°C. The slides were stained with BenchMark XT immunostainer (Ventana, Roche, Switzerland) using standard protocols, DAB Universal ultraView detection panel (Ventana, Roche, Switzerland). The following antibodies were used: HPV16 L1 (Biorbyt), MCAPL1 clone, mouse host, dilution 1:250; HPV16 L2 (Biorbyt), polyclonal, rabbit host, dilution 1:250. In order to obtain micropreparations for ICC study, the biomaterial was centrifuged in the tube for 2 minutes, at a speed of 200 rpm. BD Density Reagent was then added to the tube, centrifuged twice for 10 minutes and followed by supernatant removal. The obtained precipitate in a volume of 100 μl was placed on BD SurePath PreCoat Slides, 200 μl of reaction buffer was added and incubated for 40 min at room temperature. Next, 500 µl BD Alcohol Blend Rinse was added and incubated for 5 min. The obtained micropreparations were then dried in a thermostat for an hour at 37°C.
The IHC study was performed with a BenchMark XT immunostainer using standard protocols, using RTU antibody to Cytoactive L1 (Cytoimmun Diagnostics, Germany).
The presence of L1 or L2 protein expression in more than 10% of cells was considered a positive result. The distribution of L1 and L2 proteins was assessed using a semiquantitative method, with a score of 0 representing absence, 1 representing mild, 2 representing moderate, and 3 representing marked changes. The detection of L1 protein with the ICH study was assessed using a bicategorical method taking into account its distribution in the epithelial layer.
Statistical analysis
The sample size was calculated on the basis of the expected sensitivity and specificity of the index tests. The inclusion of 128 patients was necessary in order to achieve a power of 80% and a significance level of 0.05.
The statistical analysis of the obtained data was carried out using standard methods of mathematical and statistical processing using Statistica for Windows v.7.0 (StatSoft Inc., USA) and IBM SPSS v.22.0 programs. The Shapiro–Wilk test was used to test the normality of data distribution and the Levene's test was used for equality of variances. On the basis of these tests, a decision was made on the use of the parametric methods, including ANOVA. Nominal data were presented as absolute values (abs.) and percentage (%). Conjugation tables were usually used for independent categorical, particularly binary data, where Fisher’s exact test or Pearson’s chi-square (χ2) was most commonly used. Post-hoc analysis using Pearson’s chi-square and Benjamini–Hochberg procedure for multiple comparisons was performed when there was a statistically significant difference between groups (p<0.05). To evaluate the diagnostic accuracy of the index tests, the following parameters were used: sensitivity which is the proportion of true positives; specificity which is the proportion of true negatives; Positive Predictive Value (PPV) which is the probability of having the disease if the test result is positive; Negative Predictive Value (NPV) which is the probability of not having the disease if the test result is negative.
Results
A comparison was made of the mean values for age, weight and height of patients in the study groups (Table 1). No statistically significant differences were found (p>0.05). However, the age of patients in the HSIL group was slightly higher than the age characteristics of patients in the other groups, which is consistent with the generally accepted hypothesis of SIL development.
The analysis of the data showed that infectious diseases were statistically significantly more frequent among all somatic pathologies in the study groups (p=0.044) (Fig. 1). Thus, the frequency of HSIL detection among patients with infectious diseases was statistically significantly higher (p=0.013, Fisher–Freeman–Halton test was used, Cramer’s V coefficient = 0.334).
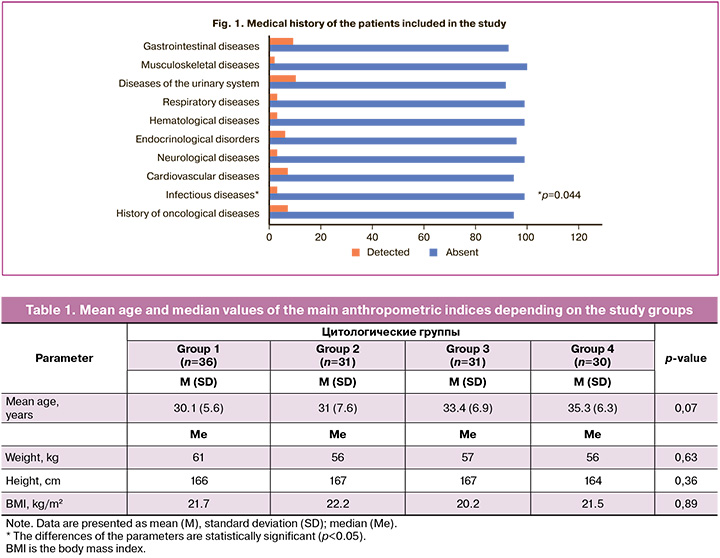
The analysis of gynecological diseases of the patients showed that statistically significant differences among the groups were obtained only for HPV-associated diseases, which occurred in almost every 2nd case and accounted for 78.1% (100/128) (p<0.001). There was an average correlation among the parameters (Cramer’s V coefficient = 0.35). Thus, there was a history of HPV-associated diseases in 50% (18/36) of the observations in the NILM group, which was statistically significantly less frequent than in the other groups (p<0.001). The rest of the parameters were not statistically significant.
The study groups were compared according to the number of HPV genotypes detected (i.e. one, two or more). It should be noted that 28.1% (36/128) of patients had multiple (two or more) HPV types of high carcinogenic risk (HCR). HPV genotype 1 was the most commonly observed among all samples analyzed, in 71.9% (92/128) of cases. The comparison of the study groups for the number of HPV types showed no statistically significant differences (p=0.39). However, multiple types of HPV HCR were more frequently detected among women in the LSIL group and accounted for 38.7% (12/31) of cases. The analysis of the frequency of different genotypes in each study group showed that DNA of HPV genotype 16 was detected most frequently and accounted for 35.2% (45/128) of all HPV genotypes detected.
Table 2 shows the results of HPV L1 marker expression using the ICC method in different cytology samples, the differences between groups were statistically significant (p<0.001). HPV L1 expression was significantly higher in LSIL group (group 3) and was 67.7% (21/31) compared to HSIL group (group 4) with 20% (6/30) and NILM group (group 1) with 19.4% (7/36) (p<0.001). In addition, the risk of detecting LSIL on cytology was increased by 2.1 times (95% CI: 1.98-10.87) among patients with positive HPV L1 expression (n=21) compared to those without this protein (n=10). Among patients with ASC-US cytology (group 2), positive and negative expression of HPV L1 protein was observed in an equal number of cases, the differences among groups were not statistically significant (p=0.14). Positive expression of HPV L1 protein was detected in 20% (6/30) of cases and negative in 80% (24/30) of cases in the HSIL group, and this was statistically significant (p=0.031). The patients with positive expression of HPV L1 (n=6) had a 4-fold lower risk of HSIL than patients without this protein (n=24) (95% CI: 0.12–0.85).
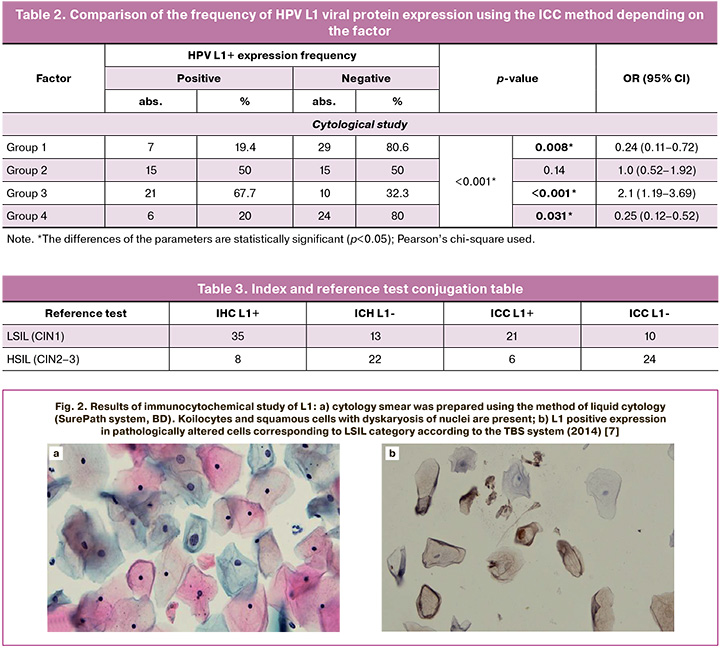
The comparison of the frequency of HPV L1 viral protein expression using the IHC method depending on the result of histological examination showed statistically significant differences (p<0.001). Positive expression of HPV L1 was significantly more frequent in the histological LSIL (CIN1) group compared to the other study groups and was 72.9% (35/48) of cases (p=0.05) (Fig. 3a, d). However, the proportion of positive HPV L1 IHC staining was 27.2% in HSIL (CIN2-3) histological specimens (Fig. 3a, c). Positive expression of this protein was observed in 15.8% and 32.1% of samples with normal histology and chronic cervicitis. The correlation between the characteristics was moderate (Cramer’s V coefficient = 0.45). The analysis of the results of HPV L2 viral protein staining intensity depending on the degree of cervical SIL (Fig. 3e, f) showed that positive expression of HPV L2 with an index of 2+ was found only in LSIL (CIN1) samples and accounted for 14.6% (7/48) of all LSIL (CIN1) cases (Figure 3b). In addition, the expression of L2 with an index of 1+ was significantly more frequently observed among specimens with LSIL (CIN1) compared to those with chronic cervicitis and HSIL (CIN2–3) (p=0.009) (Fig. 3b).
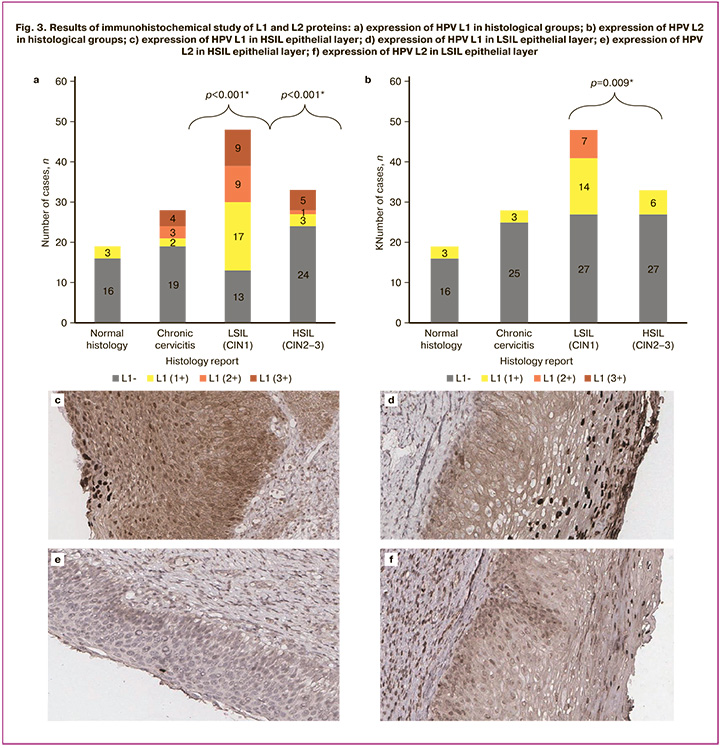
L1 expression using the ICC study was comparable to that using the IHC study and it was observed statistically significantly more often among patients with LSIL (CIN1) (p<0.001).
The present study evaluated the diagnostic significance of IHC and ICC in the use of HPV L1 and L2 proteins in the differential diagnosis of cervical SIL (Table 3). The analysis of the presented data revealed that the ICC method with HPV L1 protein had a sensitivity of 73.5% (95% CI: 60.8–83.7%) and a specificity of 84.8% (95% CI: 72.5–92.5%). This approach was also characterized by PPV of 75% (95% CI: 61.5–85.65%) and NPV of 83.8% (95% CI: 70–92.55%). However, the use of IHC study with HPV L1 protein showed a sensitivity of 72.9% (95% CI: 60.1–83.55%), specificity of 75% (95% CI: 61.5–85.6%), PPV of 63.6% (95% CI: 50–75.55%), and NPV of 83.8% (95% CI: 70–92.55%). The use of HPV L2 protein as the only marker demonstrated a sensitivity of 45.8% (95% CI: 33.1–59%), specificity of 85% (95% CI: 72.5–92.5%), PPV of 64.0% (95% CI: 48–77.5%), and NPV of 72.5% (95% CI: 59–83%). The combination of L2 and/or L1 using IHC study showed a sensitivity of 67.9% (95% CI: 58.8–77.1%) compared to the use of L2 alone, which had specificity of 80% (95% CI: 72.2–87.8%) and PPV of 64% (95% CI: 54.6–73.4%), NPV of 82.0% (95% CI: 69–90%).
Discussion
The present study evaluated the diagnostic significance of HPV L1 and L2 proteins in the differential diagnosis of cervical SIL, including cases with normal histology and chronic cervicitis using two methods (IHC and ICC) for the differential diagnosis of SIL. A further significant objective of the present study was to identify a marker that could improve the accuracy of diagnosis of lesions consistent with LSIL, which can be extremely important for the patients of reproductive age.
Currently, the HPV L1 and L2 viral proteins have been studied in separate papers, which demonstrated that their expression peaks at LSIL/CIN1, decreases significantly as koilocytes disappear from the lower and middle layers of the epithelial layer (CIN 2) and is almost completely absent at CIN 3 [8–16]. Thus, a strong and statistically significant expression of HPV L1 viral protein was observed among patients with LSIL (CIN1) compared to HSIL/CIN2-3, chronic cervicitis and normal histology using both ICC and IHC studies (p<0.001). Our results are consistent with the literature data of other studies: for example, we detected positive HPV L1 expression in LSIL samples 3 times more often than in HSIL (67.7% vs. 20%, respectively), which was higher than the results of some researchers (by 1.5 times in Mehlhorn G. et al. and 2 times in Choi Y.J. et al.) [9–12, 14]. Along with that, HPV L1 expression using the IHC method was observed statistically significantly more often among patients with LSIL (CIN1) in comparison with other patients (72.9% of cases). Thus, it can be concluded that HPV L1 expression using ICC study was comparable to the one using IHC study and was observed statistically significantly more often among patients with LSIL/CIN1 (p<0.001). The findings of the present study are consistent with the results of other studies demonstrating predominant L1 expression in lesions classified as LSIL (CIN1). Therefore, this protein can be considered as a diagnostic marker of early stages of cervical infection and it certainly has its clinical significance in the differential diagnosis of SIL [17, 18].
It should be noted that the differences between groups were not as indicative for L2 viral protein, but remained statistically significant (p=0.009). Positive expression of this marker was detected in LSIL samples in 43.5% of cases, which was 2.4 times more frequent than in HSIL (18.2% of cases). The proportion of L1-positive LSILs was lower than the proportion of L1-negative cases and it was observed 1.6 times less often compared to the frequency of HPV L1 expression among these patients; this correlates with the frequency of false-negative results for LSILs (CIN1). On the other hand, HPV L2 viral protein expression was observed in fewer specimens with morphological characteristics of HSIL (CIN2–3) and chronic cervicitis, suggesting a lower false-positive rate in these groups when HPV L2 marker was used. It is worth noting that co-expression of the two proteins was detected in 39.6% of cases in specimens with histological LSIL (CIN1), which was significantly more frequent in comparison with other groups. However, there were discrepancies in the frequency of L1 and L2 expression in the same cervical lesion in 35% of L-positive cases. It is likely that these differences may be due to the infection of the patients with more than one HPV type and there is a difference in expression related to antibody specificity in the presence of an HPV type other than HPV 16 or HPV 18. These data demonstrate that an index of the expression of HPV L2 viral protein increases the specificity of the IHC study for LSIL when the two proteins are used together, in contrast to the isolated use of HPV L1 or L2 protein.
It is important to identify the cases of positive expression of HPV viral proteins in lesions other than LSIL (CIN 1). Thus, the detection of L1 and/or L2 in normal tissue may indicate transient HPV infection. Such infection may take place when the virus is cleared by the immune system or when the virus is present in a latent form. In the latter case, the HPV genome is found in cells of the cervical squamous epithelium but no active replication occurs [19]. It should be noted that a certain ‘pool’ of samples with HSIL in the present study demonstrated both L1 and L2 expression, which was observed in a number of previous studies. Among others, Galgano T. et al. reported L1/L2 expression in 16.5% of cases [20]. In a study conducted by Yu L. et al. L1 expression was observed in 19% of cases and it was observed in 9.1% of cases in a series presented by Izadi-Mood N. et al. (2014) [21, 22]. At the same time, L1/L2 expression was not observed in any case of HSIL in studies by other authors [17, 18]. This may be due to the samples studied, which only included cases with confirmed HPV types 16 or 18. In our study, HSIL was associated with various high-risk HPV types, not only 16/18 types, and these findings are consistent with those of other authors [20].
Special attention should be paid to those cases that demonstrated the absence of L1 and/or L2 expression in the area of lesions corresponding to the histological characteristic of LSIL (CIN1). Decreased expression of L1 and L2 in HSIL (CIN2–3) is known to indicate a disruption of the viral life cycle characteristic of later stages of SIL progression [23]. In the present study, 34.4% of cases were found to be L1-negative LSIL, representing 1/3 of all LSIL cases. Our findings are consistent with the work of Yemelyanova A. et al. where more than 1/3 of LSIL cases (34.6%) were L1 negative; this may be due to the previous biomaterial collection from the cervix and colposcopy examination, which resulted in the removal of part of the surface cells from the histological specimen [17]. Evidence for this can also be found in the higher number of L1-positive specimens (72.9%) obtained by IHC study in comparison to ICC study (72.9% vs. 67.7%, respectively).
Along with this, there were 56.5% of L2-negative LSILs and 25% of lesions showing lack of L1 and L2 expression. Similar results have been previously reported in other studies, which identified L1/L2-negative LSILs in 64% of cases. Thus, the lack of L1 and/or L2 expression in LSIL (CIN1) samples may be a reflection of the localized staining of these proteins and the method of biomaterial collection (biopsy/excision/cervical conization). In other cases, it may give a false negative result due to the presence of HPV types other than those covered by the antibodies used.
In cases where L1/L2 expression was absent in the HSIL region, this absence of expression was observed not only in areas of severe dyskaryosis of epithelial cells covering the entire epithelial layer, but also in adjacent regions characterized by severe dyskaryosis in 2/3 of the epithelial layer, with preservation of koilocytes in the superficial layer. The absence of L1/L2 expression in such cases may be associated with molecular damage resulting from lesion progression, but manifesting itself before the corresponding morphological manifestation occurs. Thus, we can conclude that the use of L1 and L2 expression can significantly improve the diagnosis of LSIL, and it can exclude adjacent areas of HSIL in the verification of positive L1/L2 expression in the region of the LSIL under investigation.
In addition, regardless of the method used (IHC and ICC), data on the age-specific use of this marker were obtained. Thus, the expression level of L1 and L2 viral proteins comparably decreases in the age cohort of patients 30 years and older; the results are consistent with those of other authors [24]. These results correlate with the understanding of the HPV nature and the higher incidence of LSIL among patients younger than 30 years of age [25].
Thus, analysis of the presented data revealed that the HPV L1 protein-based ICC method demonstrated optimal sensitivity (73.5%) and reliable specificity (84.8%) against LSIL. The findings of the present study demonstrate equivalent sensitivity results with other works [9–13, 15, 22, 26–29]. Specificity in our study exceeds most of the results described previously, averaging 56.9%. This approach was also characterized by relatively high PPV of 75% and NPV of 83.8%, which indicates its acceptable ability in both detecting and excluding the diagnosis of LSIL. However, the use of IHC with HPV L1 protein showed sensitivity (72.9%) comparable to that of ICC, but its specificity was slightly lower (75%). This, in turn, resulted in a lower PPV (63.6%) and hence a lower probability of confirming the diagnosis of LSIL in case of a positive result. The use of L2 protein as the only marker showed a relatively low sensitivity (45.8%), which limits its use as a separate diagnostic tool. Despite acceptable specificity (85%), low sensitivity leads to a risk of false-negative results, especially in the early stages of the lesion. However, the combination of L2 and/or L1+ IHC showed improved sensitivity (67.9%) compared to the use of L2 alone, but specificity (80%) and PPV (64%) remained moderate. This finding suggests that the use of this combination may be more valuable as an additional test than as the main diagnostic method.
Thus, the results of this study demonstrate that the use of HPV L1 and L2 protein expression can improve the differential diagnosis of SIL. The ICC study with L1 protein showed high sensitivity (73.5%, 95% CI: 60.8–83.7%) and specificity (84.8%, 95% CI: 72.5–92.5%). This makes it a preferable method for the detection of LSIL. The combination of L1 and L2 IHC also showed improved diagnostic accuracy, which may be useful in cases of doubtful results. Further studies involving larger samples and evaluation of the diagnostic value of a combination of markers may help to optimize the diagnostic algorithm for SIL. In addition, it is important to consider the limitations of this study as well as the potential variability of L1 and L2 protein expression depending on HPV genotype and lesion stage.
Conclusion
The results of this study demonstrate that the use of HPV L1 and L2 protein expression can improve the differential diagnosis of SIL. The ICC study with L1 protein showed high sensitivity (73.5%) and specificity (84.8%), making it the preferable method for the detection of LSIL. The combination of L1 and L2 IHC also showed improved diagnostic accuracy, which may be useful in cases of doubtful results.
References
- Atla B., Uma P., Shamili M., Kumar S. Cytological patterns of cervical pap smears with histopathological correlation. Int. J. Res. Med. Sci. 2015; 3(8): 1911-6. https://dx.doi.org/10.18203/2320-6012.ijrms20150300.
- Koliopoulos G., Nyaga V.N., Santesso N., Bryant A., Martin-Hirsch P.P., Mustafa R.A. et al. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst. Rev. 2017; 8(8): CD008587. https://dx.doi.org/10.1002/14651858.CD008587.pub2.
- Singhal A., Raina R., Verma S., Verma A. Predictive accuracy of cervical cytology and colposcopy in diagnosing premalignant and malignant cervical lesions: A hospital-based study from the sub-Himalayan region of Indian subcontinent. CHRISMED Journal of Health and Research. 2019, 6(1): 39-43. https://dx.doi.org/10.4103/cjhr.cjhr_51_18.
- Wright T.C. Jr., Stoler M.H., Behrens C.M., Sharma A., Sharma K., Apple R. Interlaboratory variation in the performance of liquid-based cytology: insights from the ATHENA trial. Int. J. Cancer. 2014; 134(8): 1835-43. https://dx.doi.org/10.1002/ijc.28514.
- Kyrgiou M., Athanasiou A., Kalliala I.E.J., Paraskevaidi M., Mitra A., Martin-Hirsch P.P. et al. Obstetric outcomes after conservative treatment for cervical intraepithelial lesions and early invasive disease. Cochrane Database Syst. Rev. 2017; 11(11): CD012847. https://dx.doi.org/10.1002/14651858.CD012847.
- Kyrgiou M., Koliopoulos G., Martin-Hirsch P., Arbyn M., Prendiville W., Paraskevaidis E. Obstetric outcomes after conservative treatment for intraepithelial or early invasive cervical lesions: systematic review and meta-analysis. Lancet. 2006; 367(9509): 489-98. https://dx.doi.org/10.1016/S0140-6736(06)68181-6.
- Nayar R., Wilbur D.C. The Pap test and Bethesda 2014. Cancer Cytopathol. 2015; 123(5): 271-81. https://dx.doi.org/10.1002/cncy.21521.
- Bin H., Ruifang W., Ruizhen L., Zhihong L., Juan L., Chun W. et al. Detention of HPV L1 capsid protein and hTERC gene in screening of cervical cancer. Iranian Journal of Basic Medical Sciences. 2013; 16(6): 797-802. https://dx.doi.org/10.22038/ijbms.2013.997.
- Lee S.J., Lee A.W., Kang C.S., Park J.S., Park D.C., Ki E.Y. et al. Clinicopathological implications of human papilloma virus (HPV) L1 capsid protein immunoreactivity in HPV16-positive cervical cytology. Int. J. Med. Sci. 2013; 11(1): 80-6. https://dx.doi.org/10.7150/ijms.5585.
- Choi Y.J., Lee A., Kim T.J., Jin H.T., Seo Y.B., Park J.S. et al. E2/E6 ratio and L1 immunoreactivity as biomarkers to determine HPV16-positive high-grade squamous intraepithelial lesions (CIN2 and 3) and cervical squamous cell carcinoma. J. Gynecol. Oncol. 2018; 29(3): e38. https://dx.doi.org/10.3802/jgo.2018.29.e38.
- Norman I., Hjerpe A., Andersson S. High-risk HPV L1 capsid protein as a marker of cervical intraepithelial neoplasia in high-risk HPV-positive women with minor cytological abnormalities. Oncol. Rep. 2013; 30(2): 695-700. https://dx.doi.org/10.3892/or.2013.2538.
- Mehlhorn G., Obermann E., Negri G., Bubendorf L., Mian C., Koch M. et al. HPV L1 detection discriminates cervical precancer from transient HPV infection: a prospective international multicenter study. Mod. Pathol. 2013; 26(7): 967-74. https://dx.doi.org/10.1038/modpathol.2012.233.
- Melsheimer P., Kaul S., Dobeck S., Bastert G. Immunocytochemical detection of HPV high-risk type L1 capsid proteins in LSIL and HSIL as compared with detection of HPV L1 DNA. Acta Cytol. 2003; 47(2): 124-8. https://dx.doi.org/10.1159/000326491.
- Huang M.Z., Li H.B., Nie X.M., Wu X.Y., Jiang X.M. An analysis on the combination expression of HPV L1 capsid protein and p16INK4a in cervical lesions. Diagn. Cytopathol. 2010; 38(8): 573-8. https://dx.doi.org/10.1002/dc.21258.
- Ki E.Y., Park J.S., Lee A., Kim T.J., Jin H.T., Seo Y.B. et al. Utility of human papillomavirus L1 capsid protein and HPV test as prognostic markers for cervical intraepithelial neoplasia 2+ in women with persistent ASCUS /LSIL cervical cytology. Int. J. Med. Sci. 2019; 16(8): 1096-101. https://dx.doi.org/10.7150/ijms.31163.
- Haltas H., Bayrak R., Yenidunya S., Yildirim U. The immunohistochemical detection of P16 and HPV L1 capsid protein on cell block sections from residual PapSpin liquid-based gynecology cytology specimens as a diagnostic and prognostic tool. Eur. Rev. Med. Pharmacol. Sci. 2012; 16(11): 1588-95.
- Yemelyanova A., Gravitt P.E., Ronnett B.M., Rositch A.F., Ogurtsova A., Seidman J. et al. Immunohistochemical detection of human papillomavirus capsid proteins L1 and L2 in squamous intraepithelial lesions: potential utility in diagnosis and management. Mod. Pathol. 2013; 26(2): 268-74. https://dx.doi.org/10.1038/modpathol.2012.156.
- Negri G., Bellisano G., Zannoni G.F., Rivasi F., Kasal A., Vittadello F. et al. p16 ink4a and HPV L1 immunohistochemistry is helpful for estimating the behavior of low-grade dysplastic lesions of the cervix uteri. Am. J. Surg. Pathol. 2008; 32(11): 1715-20. https://dx.doi.org/10.1097/PAS.0b013e3181709fbf.
- Doorbar J. The papillomavirus life cycle. J. Clin. Virol. 2005; 32 Suppl 1: S7-15. https://dx.doi.org/10.1016/j.jcv.2004.12.006.
- Galgano M.T., Castle P.E., Atkins K.A., Brix W.K., Nassau S.R., Stoler M.H. Using biomarkers as objective standards in the diagnosis of cervical biopsies. Am. J. Surg. Pathol. 2010; 34(8): 1077-87. https://dx.doi.org/10.1097/PAS.0b013e3181e8b2c4.
- Izadi-Mood N., Sarmadi S., Eftekhar Z., Jahanteegh H.A., Sanii S. Immunohistochemical expression of p16 and HPV L1 capsid proteins as predictive markers in cervical lesions. Arch. Gynecol. Obstet. 2014; 289(6): 1287-92. https://dx.doi.org/10.1007/s00404-013-3124-1.
- Yu L., Wang L., Zhong J., Chen S. Diagnostic value of p16INK4A, Ki-67, and human papillomavirus L1 capsid protein immunochemical staining on cell blocks from residual liquid-based gynecologic cytology specimens. Cancer Cytopathol. 2010; 118(1): 47-55. https://dx.doi.org/10.1002/cncy.20061.
- Doorbar J., Quint W., Banks L., Bravo I.G., Stoler M., Broker T.R. et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012; 30 Suppl 5: F55-70. https://dx.doi.org/10.1016/j.vaccine.2012.06.083.
- Sarmadi S., Izadi-mood N., Pourlashkari M., Yarandi F., Sanii S. HPV L1 capsid protein expression in squamous intraepithelial lesions of cervix uteri and its relevance to disease outcome. Arch. Gynecol. Obstet. 2012; 285(3): 779-84. https://dx.doi.org/10.1007/s00404-011-2010-y.
- Viveros-Carreño D., Fernandes A., Pareja R. Updates on cervical cancer prevention. Int. J. Gynecol. Cancer. 2023; 33(3): 394-402. https://dx.doi.org/10.1136/ijgc-2022-003703.
- Byun S.W., Lee A., Kim S., Choi Y.J., Lee Y.S., Park J.S. Immunostaining of p16(INK4a)/Ki-67 and L1 capsid protein on liquid-based cytology specimens obtained from ASC-H and LSIL-H cases. Int J Med Sci. 2013; 10(12): 1602-7. https://dx.doi.org/10.7150/ijms.6526.
- Lee S.J., Lee A.W., Kim T.J., Kim J.H., Bae J.H., Lee C.W. et al. Correlation between immunocytochemistry of human papilloma virus L1 capsid protein and behavior of low-grade cervical cytology in Korean women. J. Obstet. Gynaecol. Res. 2011; 37(9): 1222-8. https://dx.doi.org/10.1111/j.1447-0756.2010.01506.x.
- Rauber D., Mehlhorn G., Fasching P.A., Beckmann M.W., Ackermann S. Prognostic significance of the detection of human papilloma virus L1 protein in smears of mild to moderate cervical intraepithelial lesions. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008; 140(2): 258-62. https://dx.doi.org/10.1016/j.ejogrb.2008.05.003.
- Xiao W., Bian M., Ma L., Liu J., Chen Y., Yang B. et al. Immunochemical analysis of human papillomavirus L1 capsid protein in liquid-based cytology samples from cervical lesions. Acta Cytol. 2010; 54(5): 661-7. https://dx.doi.org/10.1159/000325229.
Received 20.11.2024
Accepted 13.02.2025
About the Authors
Darya A. Dobrovolskaya, PhD student, V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia, +7(916)400-67-68,doc@ddoborovolskaya.ru, https://orcid.org/0000-0002-1409-9959
Guildana R. Bayramova, Dr. Med. Sci., Honored Physician of the Russian Federation, Professor at the Department of Obstetrics and Gynecology of the Department of Vocational Education, Head of Clinical Work at the Scientific Polyclinic Department, V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia, +7(926)660-48-77, bayramova@mail.ru, https://orcid.org/0000-0003-4826-661X
Alexandra V. Asaturova, Dr. Med. Sci., Head of the 1st Pathology Department, V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia, a_asaturova@oparina4.ru, https://orcid.org/0000-0001-8739-5209
Alina S. Badlaeva, PhD, Pathomorphologist at the 1st Pathoanatomical Department, V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia, a_badlaeva@oparina4.ru, https://orcid.org/0000-0001-5223-9767
Anna V. Tregubova, Pathomorphologist at the 1st Pathoanatomical Department V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, dr.a.tregubova@gmail.com, https://orcid.org/0000-0003-4601-1330



