Mitochondrial DNA expression profile in embryo culture medium in assisted reproductive technology
Objective: To investigate the feasibility of using quantitative assessment of the MT-ND1 gene copy number in culture medium as a marker of successful implantation and embryo quality. Materials and methods: We analyzed the level of MT-ND1 gene copy number in the spent culture medium as a function of embryo quality, maternal age, and the outcome of embryo transfer into the uterine cavity. Results: A total of 142 embryos were obtained, including 102 blastocysts and 40 embryos with arrested development. There was no statistically significant difference between these groups in the distribution of mtDNA copies (p=0.919). Depending on morphological characteristics, all blastocysts were divided into Group 1 (60 excellent and good embryos) and Group 2 (42 average and poor embryos). The distribution of mtDNA copies in these groups did not differ statistically significantly (p=0.082). All blastocysts were divided into groups depending on patient age, including Group 1 (<35 years, 63 embryos) and Group 2 (>35.39 years, embryos). The number of mtDNA copies was statistically significantly higher in the group of patients aged under 35 (p=0.001). Embryo transfer was performed in 24 patients. Depending on the embryo transfer outcome, patients were divided into Group 1 (negative result, 17 patients) and Group 2 (clinical pregnancy, seven patients). The distribution of mtDNA copies in these groups had no significant difference (p=0.234). Conclusion: Spent culture medium contains mtDNA derived from embryos that can be detected and analyzed by quantitative PCR. The level of mtDNA in the spent culture medium might serve as a promising marker for selecting the best embryos for transfer into the uterine cavity.Makarova N.P., Lisitsyna O.I., Nepsha O.S., Krasnyi A.M., Sadekova A.A., Nezlina A.L., Dolgushina N.V., Zingerenko B.V., Kalinina E.A.
Keywords
The main goal of fertility treatment using assisted reproductive technology (ART) is helping women become pregnant and give birth to a healthy child. Higher implantation and ongoing pregnancy rates are achieved in several ways, the main of which is selecting embryos with the highest implantation potential. The evaluation of embryos is nowadays mostly based on morphological criteria and the results of preimplantation genetic testing for aneuploidy (PGT-A). According to current studies, PGT-A increases the implantation rate per transfer, reduces the risk of pregnancy loss in the first trimester and shortens the time to pregnancy [1, 2]. At the same time, PGT-A has its disadvantages, the main of which is the invasiveness of the procedure. Direct genetic assessment of embryos through cell biopsy can decrease embryo implantation capacity.
These limitations have led to the search for non-invasive methods for evaluating human preimplantation embryos in infertility treatment programs using ART. Researchers are increasingly considering spent culture medium (collected after embryo culture and media collection) as a source of embryo DNA. It has been shown that the spent culture medium contains extracellular embryonic DNA and mitochondrial DNA (mtDNA). Analysis of spent culture medium can aid in assessing and selecting human preimplantation embryos with the highest implantation potential. Several studies have already reported a possible effective noninvasive PGT-A for the evaluation of free embryo DNA detected in the spent culture medium [3–5]. The evaluation of mtDNA isolated from the culture medium collected after embryo culture together with the morphological methods of assessing embryo quality is also promising for the selection of embryos with high potential for further development [6–12]. The results of the study suggest the possibility of non-invasive evaluation of the embryonic mtDNA content in the culture medium. Combined with embryo morphology, they can help clinical embryologists to rank embryos and selectively transfer those with the highest developmental potential.
The present study aimed to investigate the feasibility of using quantitative assessment of the MT-ND1 gene copy number in culture medium as a marker of successful implantation and embryo quality.
Materials and methods
The study was carried out in the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, V.I. Kulakov NMRC of OG&P, Ministry of Health of Russia. We analyzed 173 culture medium samples, including 142 samples containing embryos and 31 samples as a negative control without an embryo. Fifty-three married couples were included in the study. All patients underwent ovarian stimulation according to standard protocols. Thirty-six hours after ovulation initiation, the oocytes were retrieved by transvaginal follicle aspiration. Fertilization of the oocytes was performed by ICSI, after which the fertilized cells were transferred to CSCM culture medium (Irvine, Sc., USA). All steps of culture were performed in SOC (Ireland) multi-gas incubator in 25 µl oil droplets (Irvine, Sc., USA). CSCM medium (Irvine, Sc., USA) was not changed for 5–6 days of culture. On the fifth or sixth day after fertilization, the blastocysts were morphologically evaluated according to the Gardner blastocyst grading system (1999) and the RAHR guidelines for the evaluation of oocytes and embryos in the ART laboratory (2021) (Table 1).
A total of 142 embryos were evaluated, including 102 blastocysts and 40 embryos with arrested development at the stage of twinning or degenerative. Depending on morphological characteristics, all blastocysts were divided into Group 1 (60 excellent and good embryos) and Group 2 (42 fair and poor embryos). Based on the morphological evaluation, the embryos most promising for transfer were selected. We performed 24 selective embryo transfers on days 5–6 in the natural ovulation cycle. Embryos were transferred using a Wallace (Germany) or Cook (Australia) soft embryo transfer catheter. Embryos were transferred in 24 patients. Depending on the outcome of embryo transfer, patients were divided into Group 1 (negative results, 17 patients) and Group 2 (clinical pregnancy, seven patients).
In addition, all blastocysts were divided into groups categorized by patient age: Group 1 (<35) – 63 embryos; Group 2 (>35) – 39 embryos.
A drop of culture medium (31 samples) under the same culture conditions but without embryos served as a negative control. After completing embryo culture, all culture medium samples (containing and not containing embryos) were collected and cryopreserved.
The absolute copy number of Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 1 (MT-ND1) of the mtDNA gene was determined using real-time polymerase chain reaction (RT-PCR). The MT-ND1 gene encodes the NADH-ubiquinone oxidoreductase core chain 1 protein, which is a subunit of NADH dehydrogenase located in the inner mitochondrial membrane and is the largest of the five electron transfer chain complexes. For this purpose, culture medium was collected under aseptic conditions. The entire volume (10 μl) of the obtained medium was used for PCR (total volume of the mixture was 50 μl). To count mtDNA copies, we performed PCR-PV using probe primers to the mitochondrial gene of NADHdehydrogenase subunit 1 (forward primer: CCACATCTACCATCACCCTC; reverse primer: TAGAATAAATAGGAGGCCTAGGTT; probe: R6G ATC ACC GCC CCG ACC TTA GCT CTC A BHQ1). Calibration plasmid solutions (30 copies/10 μl, 125 copies/10 μl, and 250 copies/10 μl) with the appropriate amplicon cloned were used to obtain the PCR result as the absolute copy number of the NADH dehydrogenase subunit 1 gene during each run. The reaction was performed in a Real-Time PCR Detection System CFX96 amplifier (BioRad, USA) with a PCR kit (M-428, Syntol, Russia) according to the following program: 95°C – 5 min, (95°C – 10 s, 60°C – 20 s) 50 cycles. Fluorescence detection was performed in the channel corresponding to the fluorochrome probe (R6G). The obtained fluorescence data were converted into the absolute number of copies using the CFX96 BioRad amplifier software.
Statistical analysis
Statistical analysis was performed using Jamovi software. The relationship between embryo morphological quality, patient age, transfer outcomes, and culture medium mtDNA profile was evaluated. Categorical variables were presented as counts and percentages. Continuous variables were presented as the median (Me) with interquartile range (Q1; Q3). Blastocyst quality, age before and after 35 years, and embryo transfer outcome were categorical variables; mtDNA copy number was a continuous variable. To determine the statistical significance of mtDNA differences in the study groups, the nonparametric Mann–Whitney test was applied. Categorical variables were compared using Pearson's chi-square (χ2) test. The critical level of significance when testing the statistical hypothesis was considered at p <0.05.
Results
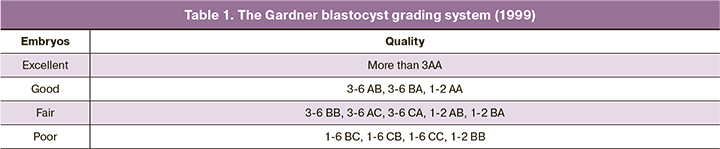
A total of 173 culture medium samples were analyzed, including 142 samples of samples collected after embryo culture from 53 married couples and 31 samples as controls. The distribution of mtDNA copies in embryo culture medium ranged from 0 to 8623 copies [median 156 (58.6; 349)]. The distribution of mtDNA copies in control culture medium ranged from 0 to 2,831 [median 7.1 (1; 14.6)], which was statistically significantly lower than in embryo culture medium, (p<0.001) (Fig. 1).
On days 5–6 of culture, 142 embryos were obtained, including 102 blastocysts of different qualities. Forty embryos were estimated to be splitting or undergoing atresia. The distribution of mtDNA copies in culture medium with embryos that reached the blastocyst stage ranged from 0 to 8623 copies [median 160 (59.4; 309)]. The distribution of mtDNA copies in culture media of embryos that were splitting or undergoing atresia was from 3.8 to 6677 copies [Me 138 (54.8; 486)], which was not significantly different (p=0.919).
Depending on the morphological grade, all blastocysts were divided into Group 1 (excellent and good) with 60 embryos and Group 2 (fair and poor) with 42 embryos. Group 1 (excellent and good blastocysts) had a median mtDNA copy distribution of 120 copies (35.5; 311); Group 2 (fair and poor) had a median mtDNA copy distribution of 198 copies (93.5; 304), which did not reach a statistically significant difference (p=0.082).
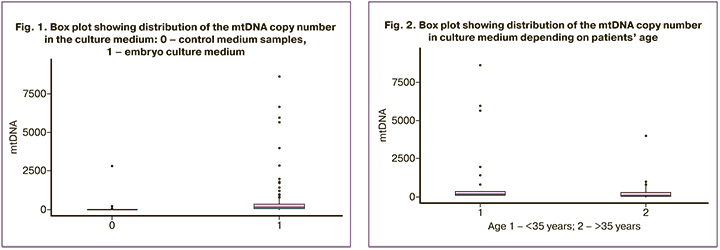
All blastocysts were divided into groups according to the age of the patients, Group 1 (<35 years) with 63 embryos and Group 2 (>35 years) with 39 embryos. The median mtDNA copy number in Group 1 (< 35) was 196 (103; 366), and in Group 2 (>35) the median mtDNA copy number was 58.5 (24.5; 294). The mtDNA copy number was statistically significantly higher in the group of the patients aged under 35 (p=0.001) (Fig. 2).
Depending on the outcome of the embryo transfer, the patients were divided into Group 1 (negative result), 17 patients, and Group 2 (clinical pregnancy), 7 patients. Group 1 (negative results) had a median mtDNA of 106 copies (39.6; 168), and Group 2 (clinical pregnancy) had a median mtDNA of 299 copies (83.6; 680), which was not statistically significantly different (p=0.234).
Differences between groups in embryo morphology (age <35 vs. >35) were assessed using Pearson's chi-square (χ2) and contingency table (Table 2). There was no statistically significant difference between the groups (p=0.093).
Besides, we separately compared mtDNA copy number depending on embryo quality (Group 1 and Group 2) among the patients aged <35 vs. >35 years. The p-value was 0.195 and 0.425, respectively, which was not statistically significant (Table 3).
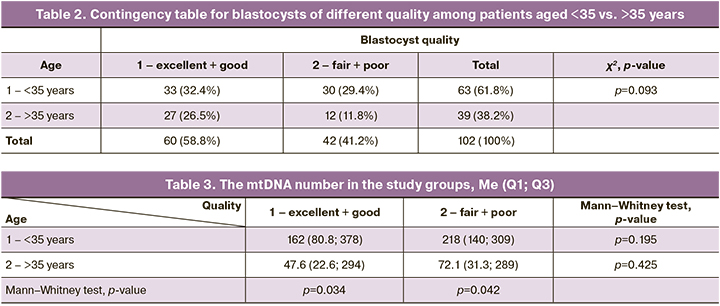
The mtDNA copy number was assessed separately depending on age (groups <35 vs. >35 years) among blastocysts of groups 1 and 2. The mtDNA copy number was statistically significantly higher in the group of patients aged <35 years: the p-value for blastocysts in groups 1 and 2 groups was 0.034 and 0.042, respectively (Table 3).
Discussion
For ART, one of the main issues still is the ability to select an embryo for transfer. Currently, morphological evaluation remains the primary method of embryo selection. However, high-grade embryos selected for transfer according to their morphological evaluation can also have aneuploid chromosomes associated with low implantation potential.
Therefore, in the field of reproductive medicine, much of the published research has focused on the development of new noninvasive methods for the evaluation of the embryo at the preimplantation stage. Areas of intensive research included microscopy techniques combined with artificial intelligence, proteomics, and metabolomic analysis of the culture medium, and analysis of cell-free DNA (in the spent culture medium and blastocyst cavity) [13–17].
Mitochondria are known to play a crucial role in cellular metabolism and function, providing cells with the necessary energy. Domestic and international authors have carried out many studies investigating mitochondria and mtDNA in follicular fluid, germ cells, embryonic cells, and blastocyst cavity fluid [18–21]. However, the results of the studies remain contradictory, and none of the methods is currently recommended for use in routine clinical practice. In addition, these methods involve invasive intervention. The analysis of cell-free DNA in the culture medium, including mtDNA, is considered a promising and innovative method of preimplantation embryo evaluation. Researchers suggest a role for mtDNA as a potential marker of implantation success. However, studies investigating mtDNA in culture medium are lacking in the world literature.
In our study, we examined the relationship between embryo morphological quality, patient age, transfer results, and mtDNA profile in culture medium. Additionally, the numbers of mtDNA copies in culture medium without embryos and under the same conditions were studied as a control.
The findings confirm that the culture medium is a source of mtDNA that can be detected and analyzed by quantitative PCR. This agrees with the data of other researchers, claiming that mtDNA can be detected in culture medium by PCR in 99% of cases [9].
According to our findings, the number of mtDNA copies in culture medium is significantly higher in the group of patients under 35 years old. It should be noted that a statistically significant difference was observed when comparing all blastocysts (in maternal age groups <35 and >35 years) and when separately comparing blastocysts of different qualities. These results are consistent with the findings of the 2019 study by S. Stigliani et al. [11]. In contrast, according to other studies, the number of mtDNA is higher in patients of older reproductive age [9, 12]. Many authors noted the absence of any relationship between mtDNA levels in the culture medium and maternal age [6, 8].
We also observed that the mtDNA copy number in the culture medium was not associated with the level of embryo development on days 5-6, with the morphological evaluation of the blastocyst and successful implantation rate. Similar observations were reported by other researchers [6, 8]. In addition, different results are described in the literature.
A team of researchers led by S. Stigliani in 2013 concluded that a higher concentration of mtDNA in the culture medium on days 2–3 of culture was associated with the degree of embryo fragmentation and with the older reproductive age of patients (35 years and older) [9]. In a more recent 2014 study, the same authors analyzed 605 samples of culture medium on day 3 and obtained the opposite results: a high concentration of mtDNA was a predictor of successful blastulation of good quality embryos with a low (mild) degree of fragmentation. Furthermore, a high concentration of mtDNA was associated with a successful implantation rate in 3-day embryo transfer [10]. The authors analyzed not only the copy number, but also the ratio of mtDNA to genomic DNA in the culture medium. In 2019, the researchers complemented their study and attempted to develop a combined approach to embryo selection [11]. The authors emphasized that mtDNA levels were significantly higher for embryos that reached the blastocyst stage compared to those with arrested development. Furthermore, the amount of mtDNA in the culture medium was higher among women under 35 years of age. Thus, the researchers concluded that the mtDNA/genomic DNA ratio at day 3 (threshold value ≥184) combined with morphological evaluation improves the effectiveness of blastocyst prediction, regardless of patient and cycle characteristics (sensitivity, 95%; specificity, 65%).
Another team of authors determined that the amount of mtDNA in the spent culture medium has a positive correlation with the amount of mtDNA in the blastocyst cells (the whole blastocyst material was examined), with maternal age and embryo quality [12].
Studies have shown that the concentration of mtDNA in the culture medium is significantly higher than that of genomic DNA [9, 18]. Some researchers note that the mtDNA copy number in the culture medium is higher on day 5 compared to day 3 of culture [7]. Other authors emphasize the absence of a relationship between the level of mtDNA in culture medium and ploidy of the embryo [6, 12]. According to Kobayashi M. et al., mtDNA copy number in culture medium is higher for blastocysts experiencing blastocyst collapse and has a positive correlation with their number [8]. Transfer of blastocysts experiencing blastocyst collapse episodes is known to be associated with a decrease in the successful implantation from 48.5 to 35.1% [22]. The same authors note that the time between blastocyst initiation and the formation of a complete blastocyst has a significant positive correlation with the amount of mtDNA in the culture medium [8].
Taking into consideration our data as well as the results of other similar studies, the issue of the possible use of mtDNA copy number in the culture medium as a noninvasive marker of embryo selection for transfer into the uterine cavity remains open and requires further research.
The research evidence published to date in the global literature is extremely contradictory, which can be attributed to several factors. The main ones are the small number of studies on mtDNA in culture medium, small sample sizes, different study designs, different methods of mtDNA analysis (PCR, NGS) and the choice of different fragments for mtDNA detection. In addition, some studies were performed in one clinic (as in our case) and some in several clinics. Such factors as conditions and duration of embryo culture and the type of culture medium and time (days) of its collection could have influenced the obtained results. Among other factors, it should be noted that the morphological evaluation of embryos is performed by an embryologist and is subjective in nature, which can also affect the final results of the study.
At the same time, the presence from isolated results of control culture medium containing high levels of mtDNA indicates possible external contamination during the collection, transportation, or analysis of the material. Common causes of contamination can include protein components of the culture medium, maternal or paternal contamination (by cumulus cells, polar corpora, or spermatozoa), and external contamination during the culture period [18].
Our study was carried out as a pilot project and had a small sample size, which probably influenced the result. Nevertheless, this work shows that the mtDNA copy number in embryo culture medium is higher in patients under 35 years compared to those of older reproductive age. The mtDNA copy number is not associated with the degree of embryo development at day 5–6 of culture, the morphological evaluation of the blastocyst, or the successful implantation rate.
Additional large, appropriately designed and powered multicenter studies are required. They should use unified mtDNA analysis, several variants of control (blank culture medium on days 0, 3, and 5 of culture), filters during material handling, and several fragments (primers) for mtDNA detection.
Conclusion
Spent culture medium contains mtDNA derived from embryos that can be detected and analyzed by quantitative PCR. The level of mtDNA in the spent culture medium might serve as a promising marker for selecting the best embryos for transfer into the uterine cavity. More research is required to develop a unified approach to mtDNA detection and culture protocols, and to determine the feasibility of their use in clinical practice.
References
1. Shi W.H., Jiang Z.R., Zhou Z.Y., Ye M.J., Qin N.X., Huang H.F. et al. Different strategies of preimplantation genetic testing for aneuploidies in women of advanced maternal age: A systematic review and meta-analysis. J. Clin. Med. 2021; 10(17): 3895. https://dx.doi.org/10.3390/jcm10173895.
2. Bhatt S.J., Marchetto N.M., Roy J., Morelli S.S., McGovern P.G. Pregnancy outcomes following in vitro fertilization frozen embryo transfer (IVF-FET) with or without preimplantation genetic testing for aneuploidy (PGT-A) in women with recurrent pregnancy loss (RPL): a SART-CORS study. Hum. Reprod. 2021; 36(8): 2339-44. https://dx.doi.org/10.1093/humrep/deab117.
3. Brouillet S., Martinez G., Coutton C., Hamamah S. Is cell-free DNA in spent embryo culture medium an alternative to embryo biopsy for preimplantation genetic testing? A systematic review. Reprod. Biomed. Online. 2020; 40(6): 77996. https://dx.doi.org/10.1016/j.rbmo.2020.02.002.
4. Chen L., Sun Q., Xu J., Fu H., Liu Y., Yaxin Yao Y. et al. A non-invasive chromosome screening strategy for prioritizing in vitro fertilization embryos for implantation. Front. Cell Dev. Biol. 2021; 9: 708322. https://dx.doi.org/10.3389/fcell.2021.708322.
5. Rubio C., Racowsky C., Barad D.H., Scott R.T., Simon C. Noninvasive preimplantation genetic testing for aneuploidy in spent culture medium as a substitute for trophectoderm biopsy. Fertil. Steril. 2021; 115(4): 841-9. https:// dx.doi.org/10.1016/j.fertnstert.2021.02.045.
6. Zhang X., Sun Y., Dong X., Zhou J., Sun F., Han T. et al. Mitochondrial DNA and genomic DNA ratio in embryo culture medium is not a reliable predictor for in vitro fertilization outcome. Sci. Rep. 2019; 9(1): 5378. https://dx.doi.org/10.1038/s41598-019-41801-1.
7. Hammond E., McGillivray B., Wicker S., Peek J.C., Shelling A.N., Stone P. et al. Characterizing nuclear and mitochondrial DNA in spent embryo culture media: genetic contamination identified. Fertil. Steril. 2017; 107(1): 220-8. e5. https://dx.doi.org/10.1016/j.fertnstert.2016.10.015.
8. Kobayashi M., Kobayashi J., Shirasuna K., Iwata H. Abundance of cell-free mitochondrial DNA in spent culture medium associated with morphokinetics and blastocyst collapse of expanded blastocysts. Reprod. Med. Biol. 2020; 19(4): 404-14. https://dx.doi.org/10.1002/rmb2.12344.
9. Stigliani S., Anserini P., Venturini P., Scaruffi P. Mitochondrial DNA content in embryo culture medium is significantly associated with human embryo fragmentation. Hum. Reprod. 2013; 28(10): 2652-60. https://dx.doi.org/10.1093/humrep/det314.
10. Stigliani S., Persico L., Lagazio C., Anserini P., Venturini P., Scaruffi P. Mitochondrial DNA in Day 3 embryo culture medium is a novel, non- invasive biomarker of blastocyst potential and implantation outcome. Mol. Hum. Reprod. 2014; 20(12): 1238-46. https://dx.doi.org/10.1093/molehr/ gau086.
11. Stigliani S., Orlando G., Massarotti C., Casciano I., Bovis F., Anserini P. et al. Non-invasive mitochondrial DNA quantification on Day 3 predicts blastocyst development: a prospective, blinded, multi-centric study. Mol. Hum. Reprod. 2019; 25(9): 527-37. https://dx.doi.org/10.1093/molehr/ gaz032.
12. Zhang J., Xia H., Chen H., Yao C., Feng L., Song X., Bai X. Less-invasive chromosome screening of embryos and embryo assessment by genetic studies of DNA in embryo culture medium. J. Assist. Reprod. Genet. 2019; 36(12): 250513. https://dx.doi.org/10.1007/s10815-019-01603-w.
13. Zmuidinaite R., Sharara F.I., Iles R.K. Current advancements in noninvasive profiling of the embryo culture media secretome. Int. J. Mol. Sci. 2021; 22(5): 2513. https://dx.doi.org/10.3390/ijms22052513.
14. Валиахметова Э.З., Кулакова Е.В., Скибина Ю.С., Грязнов А.Ю., Сысоева А.П., Макарова Н.П., Калинина Е.А. Неинвазивное тестирование преимплантационных эмбрионов человека in vitro как способ прогнозирования исходов программ экстракорпорального оплодотво-рения. Акушерство и гинекология. 2021; 5: 5-16. [Valiakhmetova E.Z., Kulakova E.V., Skibina Yu.S., Gryaznov A.Yu., Sysoeva A.P., Makarova N.P., Kalinina E.A. Non-invasive testing of human preimplantation embryos in vitro as a way to predict the outcomes of in vitro fertilization programs. Obstetrics and Gynecology. 2021; 5: 5-16. (in Russian)]. https://dx.doi.org/10.18565/ aig.2021.5.5-16.
15. Ярыгина С.А., Смольникова В.Ю., Калинина Е.А., Эльдаров Ч.М., Гамисония А.М., Макарова Н.П., Бобров М.Ю. Анализ метаболитов в различных средах культивирования эмбрионов человека. Акушерство и гинекология. 2020; 11: 114-23. [Yarygina S.A., Smolnikova V.Yu., Kalinina E.A., Eldarov Ch.M., Gamisonia A.M., Makarova N.P., Bobrov M.Yu. Analysis of human embryo culture medium metabolites. Obstetrics and Gynecology. 2020; 11: 114-23. (in Russian)]. https://dx.doi.org/10.18565/aig.2020.11.114-123.
16. Сысоева А.П., Макарова Н.П., Калинина Е.А., Скибина Ю.С., Занишевская А.А., Янчук Н.О., Грязнов А.Ю. Повышение эффективности вспомогательных репродуктивных технологий с помощью искусственного интеллекта и машинного обучения на эмбриологическом этапе. Акушерство и гинекология. 2020; 7: 28-36. [Sysoeva A.P., Makarova N.P., Kalinina E.A., Skibina Yu.S., Zanishevskaya A.A., Yanchuk N.O., Gryaznov A.Yu. Enhancing the efficiency of assisted reproductive technologies using artificial intelligence and machine learning at the embryological stage. Obstetrics and Gynecology. 2020; 7: 28-36. (in Russian)]. https://dx.doi.org/10.18565/aig.2020.7.28-36.
17. Тимофеева А.В., Калинина Е.А., Драпкина Ю.С., Чаговец В.В., Макарова Н.П., Сухих Г.Т. Оценка качества эмбриона по профилю экспрессии малых некодирующих РНК в культуральной среде эмбриона в программах ВРТ. Акушерство и гинекология. 2019; 6: 79-86. [Timofeeva A.V., Kalinina E.A., Drapkina Yu.S., Chagovets V.V., Makarova N.P., Sukhikh G.T. Embryo quality assessment by the small noncoding RNA expression profile in an embryo culture medium in assisted reproductive technology programs. Obstetrics and Gynecology. 2019; 6: 79-86. (in Russian)]. https://dx.doi.org/10.18565/ aig.2019.6.79-86.
18. Hammond E, Shelling A, Cree L. Nuclear and mitochondrial DNA in blastocoele fluid and embryo culture medium: evidence and potential clinical use. Hum. Reprod. 2016; 31(8): 1653-61. https://dx.doi.org/10.1093/humrep/dew132.
19. Королькова А.И., Мишиева Н.Г., Мартазанова Б.А., Бурменская О.В., Екимов А.Н., Трофимов Д.Ю., Веюкова М.А., Кириллова А.О., Абубакирова А.Н. Повышение эффективности программ ЭКО на основании определения копийности митохондриальной ДНК в трофэктодерме эмбрионов. Акушерство и гинекология. 2019; 3: 98-104. [Korolkova A.I., Mishieva N.G., Martazanova B.A., Bourmenskaya O.V., Ekimov A.N., Trofimov D.Yu., Veyukova M.A., Kirillova A.O., Abubakirov A.N. Increasing the effectiveness of IVF programs by determining mitochondrial DNA copy number in embryonic trophectoderm. Obstetrics and Gynecology. 2019; 3: 98-104. (in Russian)]. https://dx.doi.org/10.18565/aig.2019.3.98-104.
20. Перминова С.Г., Митюрина Е.В., Селимова Ф.Н., Бурменская О.В., Козырина Н.В., Кравченко А.В. Копийность митохондриальной ДНК в эякуляте ВИЧ-инфицированных мужчин, принимающих антиретровирусную терапию. Акушерство и гинекология. 2020; 4: 120-6. [Perminova S.G., Mityurina E.V., Selimova F.N., Burmenskaya O.V., Kozyrina N.V., Kravchenko A.V. Mitochondrial DNA copy number in sperm of HIV-infected men receiving antiretroviral therapy. Obstetrics and Gynecology. 2020; 4: 120-6. (in Russian)]. https://dx.doi.org/10.18565/aig.2020.3.120-126.
21. Непша О.С., Кулакова Е.В., Екимов А.Н., Драпкина Ю.С., Макарова Н.П., Краевая Е.Е., Калинина Е.А. Использование митохондриальной ДНК эмбрионов в качестве предиктора эффективности программ вспомогательных репродуктивных технологий. Акушерство и гинекология. 2021; 11: 125-34. [Nepsha O.S., Kulakova E.V., Ekimov A.N., Drapkina Yu.S., Makarova N.P., Kraevaya E.E., Kalinina E.A. Value of embryonic mitochondrial DNA in predicting the effectiveness of assisted reproductive technologies. Obstetrics and Gynecology. 2021; 11: 125-34. (in Russian)].
22. Marcos J., Perez-Albala S., Mifsud A., Malla M., Landeras J., Meseguer M. Collapse of blastocysts is strongly related to lower implantation success: a timelapse study. Hum. Reprod. 2015; 30(11): 2501-8. https://dx.doi.org/10.1093/ humrep/dev216.
Received 18.01.2022
Accepted 24.02.2022
About the Authors
Nataliya P. Makarova, Dr. Bio. Sci., Dr. Bio. Sci., Leading Researcher at at the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility,Academician V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, np_makarova@oparina4.ru, https://orcid.org/0000-0003-1396-7272, 4 Oparina str., Moscow, 117997, Russia.
Olga I. Lisitsyna, Postgraduate Student, Academician V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health
of the Russian Federation, o_yazykova@inbox.ru, https://orcid.org/0000-0002-7775-3508, 4 Oparina str., Moscow, 117997, Russia.
Oksana S. Nepsha, Ph.D., Researcher at the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, Academician V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, o_nepsha@oparina4.ru, https://orcid.org/0000-0002-9988-2810, 4 Oparina str., Moscow, 117997, Russia.
Aleksey M. Krasnyi, Ph.D. (Bio. Sci.), Head of the Cytology Laboratory, Academician V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, alexred@list.ru, https://orcid.org/0000-0001-7883-2702, 4 Oparina str., Moscow, 117997, Russia.
Alsu A. Sadekova, PhD (Bio), Researcher of the Cytology Laboratory, Academician V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, sialsad@gmail.com, https://orcid.org/0000-0003-4726-7477, 4 Oparina str., Moscow, 117997, Russia.
Alexandra L. Nezlina, Junior Researcher at the Cytology Laboratory, Academician V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, nezlina@vk.com, 4 Oparina str., Moscow, 117997, Russia.
Nataliya V. Dolgushina, Dr. Med. Sci., Professor, Deputy Director - Head of the Department of Research Administration, Academician V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, n_dolgushina@oparina4.ru, https://orcid.org/0000-0003-1116-138X, 4 Oparina str., Moscow, 117997, Russia.
Boris V. Zingerenko, Researcher at the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, Academician V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, regniz@mail.ru, https://orcid.org/0000-0002-8784-5502, 4 Oparina str., Moscow, 117997, Russia.
Elena A. Kalinina, Dr. Med. Sci., Professor, Head of the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, Academician V.I. Kulakov National
Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of the Russian Federation, e_kalinina@oparina4.ru, https://orcid.org/0000-0002-8922-2878, 4 Oparina str., Moscow, 117997, Russia.
Authors' contributions: Makarova N.P., Nepsha O.S., Lisitsyna O.I., Dolgushina N.V - conception and design of the study;
Lisitsyna O.I., Nepsha O.S., Makarova N.P., Dolgushina N.V. - manuscript drafting and editing; Lisitsyna O.I, Nepsha O.S. - statistical analysis; Zingerenko B.V. - collection of biological material; Krasny A.M., Nezlina A.L., Sadekova A.A. - laboratory stage (mtDNA analysis by PCR); Dolgushina N.V., Kalinina E.A. - manuscript approval.
Conflicts of interest: The authors have no conflicts of interest to declare.
Funding: There was no funding for this study.
Ethical Approval: The study was reviewed and approved by the Research Ethics Committee of the V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia.
Patient Consent for Publication: All patients provided informed consent for the publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator.
For citation: Makarova N.P., Lisitsyna O.I., Nepsha O.S., Krasnyi A.M., Sadekova A.A., Nezlina A.L., Dolgushina N.V., Zingerenko B.V., Kalinina E.A. Mitochondrial DNA expression profile in embryo culture medium in assisted reproductive technology.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2022; 3: 89-96 (in Russian)
https://dx.doi.org/10.18565/aig.2022.3.89-96



