Experience of using high-throughput sequencing (NGS) for noninvasive prenatal screening of fetal aneuploidy at the D.O. Ott Research Institute of Obstetrics, Gynecology and Reproduction
Objective: To evaluate the clinical efficiency of noninvasive prenatal screening of fetal aneuploidies in the mother’s blood conducted at the D.O. Ott Research Institute of Obstetrics, Gynecology and Reproduction from December 2019 to April 2022 in comparison with first trimester combined screening.Tarasenko O.A., Vashukova E.S., Kozyulina P.Yu., Morshneva A.V., Maltseva A.R., Pachulia O.V., Talantova O.E., Koroteev A.L., Ivashchenko T.E., Bespalova O.N., Kogan I.Yu., Baranov V.S., Glotov A.S.
Material and methods: A total of 4272 blood samples obtained from pregnant women were analyzed. Noninvasive prenatal screening of fetal aneuploidies was performed by means of the analysis of extracellular DNA (eDNA) of the fetus in the mother’s blood by high-throughput sequencing (NGS).
Results: A high risk of fetal aneuploidy was noted in 173/4272 cases (4.05% of all studies). The results were confirmed by invasive diagnostic methods in 138 cases. Among them, the most common disorders are trisomy 21 (Down syndrome) – 74.63% of cases, trisomy 18 (Edwards syndrome) – 7.97%, trisomy 13 (Patau syndrome) – 6.52%; these results are consistent with the findings obtained in the Russian and foreign practice. The prognostic value of detecting trisomy 21, 18, and 13 is significantly higher in comparison with first trimester combined screening. There is a high percentage of fetuses with abnormalities (2.41%) in women of the so-called fetal concern group; their percentage is equal to that (2.46%) in pregnant women of “intermediate risk” according to the results of early prenatal screening.
Conclusion: The results of the study show the high clinical efficacy of noninvasive prenatal screening of fetal aneuploidy by NGS method. In case of detecting a high risk of fetal chromosomal pathology, it is necessary to refer the patient to a geneticist for consultation and for solving the issue of invasive prenatal diagnosis as well.
Keywords
Prenatal diagnosis of congenital and hereditary diseases is one of the priority areas of medical genetics. Chromosomal diseases are known to occupy one of the leading places among human genetic diseases. According to cytogenetic studies, the incidence of chromosomal pathology among newborns is 0.6–1.0% [1].
Numerical chromosomal abnormalities are the most common variant of chromosomal pathology. However, relatively few of them are compatible with postnatal development and result in chromosomal diseases [1]. The most severe and widespread chromosomal abnormalities include trisomy 21 (Down syndrome) [2], trisomy 18 (Edwards syndrome), trisomy 13 (Patau syndrome) [3].
According to the Order of the Ministry of Health of the Russian Federation dated October 20, 2020 No. 1130n “On approval of the procedure for providing medical care in the profile Obstetrics and Gynecology”, the first stage of prenatal diagnosis is combined early prenatal screening (EPS), which includes ultrasound examination, identification of serum markers of maternal blood and evaluation of individual risk for chromosomal abnormalities. EPS is performed in the first trimester of pregnancy at 11–14 weeks gestation and detects up to 90% (84% on average in Russia) of fetuses with common aneuploidies [4–6].
It should be noted that conventional prenatal screening is based on indirect methods of fetal condition assessment (ultrasound and biochemical screening), therefore, it is impossible and will be impossible in the future to achieve a higher effectiveness of the detection of fetal genetic pathology.
In turn, noninvasive prenatal DNA screening (NIPT, NIPS) is based on direct examination of fetal DNA [7]. The rate of false positive results in this type of screening is lower than in standard prenatal screening [8]. However, despite the high levels of sensitivity and specificity, NIPS requires diagnostic confirmation, since DNA is analyzed during this procedure. DNA enters the maternal bloodstream from cytotrophoblast cells, which are the material of the placenta, not the fetus [5].
Nowadays, the place of NIPS in prenatal diagnostics is a subject for discussion. According to the current clinical guidelines “Normal Pregnancy” (approved by the Ministry of Health of Russia in 2020), a patient may additionally be offered noninvasive screening after 10 weeks gestation in order to exclude fetal aneuploidy. However, the total introduction of NIPS for all pregnant women is premature for a number of reasons including the high cost of the test, labor intensity, duration, as well as its screening characteristics. At the same time, it is necessary to use this technology if a woman wishes or if a patient is in the “intermediate risk group” after performing EPS since more than 10% of patients with fetal trisomy are not detected by conventional EPS. NIPS can also be administered in a high-risk group of patients in case of fear or contraindications to invasive prenatal diagnosis [6]. Thus, NIPS can be considered as a screening procedure (for the presence of aneuploidy in the fetus) on the one hand, and additional study to EPS, on the other hand. However, in the case of a positive result, the diagnosis must be confirmed using invasive techniques [7, 9].
Different variants of NIPS can be used to screen chromosomal abnormalities by extracellular DNA of the fetus. The first option is a genome–wide study. In this test, the technology of genome-wide high-throughput DNA sequencing (NGS) is used: DNA which circulates in the blood plasma of a pregnant woman, including the genomes of the mother and fetus with low coverage (0.3–0.5X), is sequenced and the ratio of copies of DNA fragments of various fetal chromosomes to those of the mother is calculated. The test makes it possible to detect aneuploidies in all chromosomes, as well as to analyze partial trisomy and monosomy (deletions and duplications), changes in the form of a variation in the number of copies of DNA (Copy number variation, CNV) that are not detected by standard screening [8, 10, 11].
The second option is a targeted test. It can be performed using NGS technology (with high coverage – 200–1000X) and others, for example, microchips, real–time polymerase chain reaction, ring rolling technology, etc. [12]. Screening for aneuploidies of five chromosomes (21, 13, 18, X, Y) and a number of deletion syndromes is usually carried out as part of this test.
In our study, we performed NIPS for all the studied samples in a genome-wide format.
The aim of the study was to evaluate the clinical efficiency of noninvasive prenatal screening of fetal aneuploidies in the mother’s blood which was conducted at the D.O. Ott Research Institute of Obstetrics, Gynecology and Reproduction from December 2019 to April 2022 in comparison with first trimester combined screening.
Materials and methods
A total of 4272 noninvasive prenatal studies were conducted in the period from December 2019 to April 2022; women with a singleton pregnancy at ≥ 9 weeks gestation from 16 to 49 years old (average age 34.34 (4.49) years) participated in them.
All participants gave their informed consent; they were also informed about the possibilities and limitations of NIPS, as well as the need for an invasive screening in case of a positive result. The study was approved by the Ethical Review Board of the D.O. Ott Research Institute of Obstetrics, Gynecology and Reproduction, St. Petersburg, Russia (Protocol No.130 – 16/07/2020) and it was carried out in accordance with the Helsinki Declaration.
Venous Blood was collected in test tubes containing 0.5 M EDTA, as well as in Cell-Free DNA BCT (Streck, USA) tubes containing a stabilizer of nucleated blood cells. Blood in Cell-Free DNA BCT tubes can be stored for up to two weeks at room temperature; thus, samples can be transported from any region of Russia. If blood was collected in 0.5M EDTA tubes, then plasma was isolated no later than 4 hours after blood collection. In case of collecting biomaterial in a Cell-Free DNA BCT tube, plasma was isolated at the time of sample obtaining after its transportation.
DNA isolation, library preparation, genome-wide next generation sequencing (instructions with modifications) and bioinformatic processing of the obtained results were carried out in accordance with the patented protocol (patent RU 02712175 C1 20200124).
Statistical analysis
Statistical processing of the obtained results was carried out using the Microsoft Excel 2002 software. The significance of frequency differences was determined using the Pearson chi-squared test (2) according to the standard formula. Mean value and standard deviation were used to characterize the level of fetal DNA fraction. The results of the correlation analysis of the dependence of the fetal fraction level on the woman’s body mass index and gestational age were obtained using the Graph Pad Prism program.
In order to learn the opinion of patients about NIPS, we conducted a survey of 32 women. The sample was formed randomly. The questionnaires were processed using the Microsoft Excel 2002 tables.
Results
NIPS was introduced into practice at the D.O. Ott Research Institute of Obstetrics, Gynecology and Reproduction in December 2019. The test was developed in cooperation with LLC NIPT, St. Petersburg, Russia. The technology was used in two ways: genome-wide (aneuploidy was determined for all chromosomes) and targeted (aneuploidy was determined for chromosomes 13, 18, 21, X, Y). The test was proven at the D.O. Ott Research Institute of Obstetrics, Gynecology and Reproduction; it was verified at Family Planning and Reproduction Center (Moscow), Saint-Petersburg State Pediatric Medical University, Stavropol Regional Clinical Perinatal Center No. 1, Orenburg Regional Clinical Hospital No. 2.
This study presents the results of analysis of cell-free fetal DNA (cffDNA) in the blood of 4272 pregnant women. All pregnant women underwent genome-wide screening of extracellular DNA (Table 1).
The average age of all women was 34.34 years (16–49 years); there were 2182 (51.08%) women younger than 35 years, and 2090 (48.92%) women aged 35 years and older at the moment when the study was conducted. The largest number of women who took part in the study were in the following age groups: 1276/4272 (29.86%) from 30 to 34 years old and 1344/4272 (31.46%) from 35 to 39 years old. The smallest group consisted of patients under 20 years of age, namely 18/4272 (0.42%).
The body mass index of pregnant women ranged from 15.94 to 51.42 kg/m2 (the mean value is 24.29 kg/m2). The largest group consisted of women with normal body weight (2584/4272 (60.49%)). However, about a third of the patients were obese or overweight (1495/4272 (34.99%)).
The gestation period ranged from 8.5 to 30 weeks (the mean value was 14.8 weeks). The largest number of pregnant women had gestation between 12 and 16 weeks (2489/4272 (58.26%)). A small number of women had the test earlier than 10 weeks (54/4272 (1.27%)). There was a significant number of women who agreed to take the test at 22 weeks gestation and later (121/4272 (2.82%)).
Patients became pregnant after assisted reproduction in 364/4272 cases (8.52%) (pregnancy occurred as a result of IVF), including 75 cases (1.75%) when a donor egg was used.
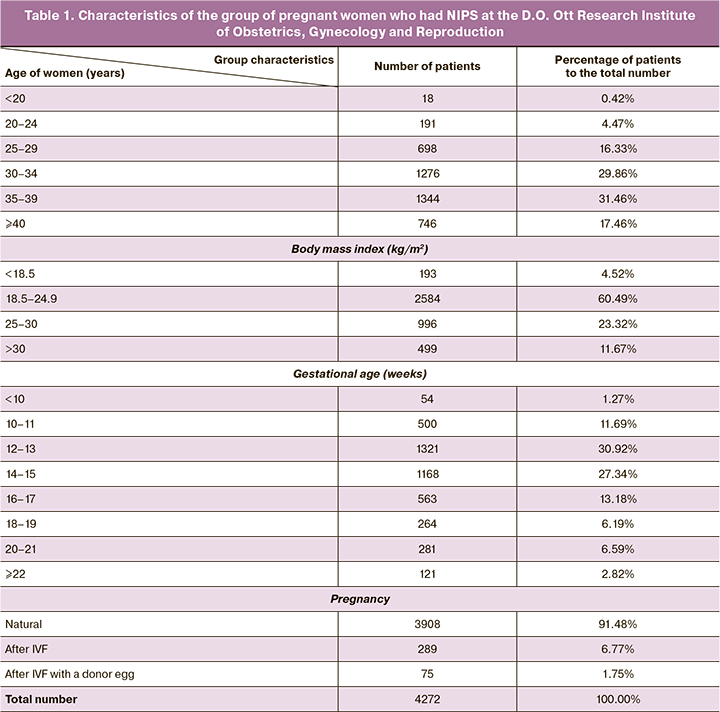
In general, noninvasive prenatal screening was performed in 99.6% of cases. Low risk of chromosomal abnormalities was found in 4080/4272 (95.6%) patients, a high risk of chromosomal abnormalities was found in 173/4272 (4.05%), and the result of the screening was not obtained in 19/4272 (0.44%) cases (Fig. 1). Trisomy 21 prevailed among the detected abnormalities (103/173 cases, 60%); trisomy 18, trisomy 13 and sex chromosomes were detected in 13/173 (8%), 12/173 (7%) and 11/173 (6%) cases, respectively. Rare trisomies, so-called “accidental findings”, accounted for 34/173 (19%) cases.
In order to analyze the data in detail, we studied the following indicators:
A) General characteristics of NIPS
- frequency of repeated blood sampling for examination;
- the level of fetal fraction (FF) in pregnant women;
B) The frequency and type of chromosomal pathologies detected by NIPS
- representation of chromosomal abnormalities depending on medical indications;
- the effectiveness of NIPS for detecting chromosomal pathologies.
C) Awareness of pregnant women about NIPS and satisfaction with the provided service.
General characteristics of the test
Cases of repeated blood sampling for research
NIPS is an expensive service and its use largely depends on the decision of a pregnant woman, so the quality of this study is of great scientific and practical importance. The quality of the test can be assessed by a number of parameters, such as the number of cases of repeated sampling of biomaterial, the number of cases with absent test results after repeated testing, FF level, maternal DNA content, and others [13–16].
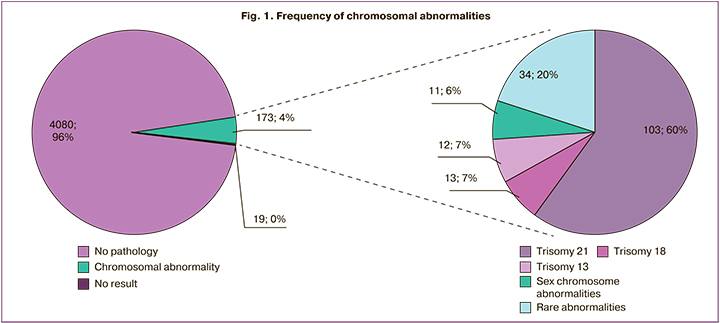
In our study, blood was sampled repeatedly in 116 (2.7%) cases out of 4272. It was done due to the following reasons: low FF content (n=39), hemolysis (n=42), questionable results in determining the gender of the fetus (n=18) and aneuploidy (n=9), samples did not pass quality control (n=8). The result of NIPS was obtained in 97 cases after repeated blood sampling. Thus, the final conclusion based on the results of the study was not made only in 19/4272 (0.44%) cases. We were not able to determine the gender of the fetus in ten of them, five pregnant women refused to repeat the study and the FF level in the sample remained below 4% in four pregnant women (the number of results which were not obtained decreases to 0.33% (14/4272) when women who refused to repeat the study are excluded from the sample).
The obtained percentage of cases (0.33%) without test results after repeated examination was significantly lower than in the previous studies published by Russian colleagues [17, 18] and can be compared with the published data of a number of foreign researchers [19].
It should be noted that along with an increase in the number of conducted studies there is a tendency to a decrease in the number of samples, which require repeated sampling due to the low FF level, questionable results in determining the gender of the fetus, poor quality of samples, as well as samples without results obtained after repeated examination. Figure 2 shows this dynamics (relative number of cases), which was revealed during the evaluation of 2345 and 4272 studies in 2021 and 2022, respectively.
FF analysis
In order to obtain a reliable result of DNA screening, it is necessary to have an FF level of at least 4% [20]. In women who took part in our study, it ranged from 4.04% to 39.13% (the average value is 11.82%)) and it corresponds to similar data obtained by other Russian researchers [13, 14].
However, it was not possible to achieve the desired parameter of FF in some cases during the initial study and it was necessary to take the patient’s blood again. In our study, 39 out of 4272 samples showed low FF (less than 4% of all extracellular DNA); among them 26/39 (66.7%) samples belonged to women with a high (≥30 kg/m2) body mass index and 13/39 (33.3%) samples belonged to women without overweight. Repeated blood sampling allowed us to obtain a result in 35 out of 39 cases; the result was not obtained in 4 cases (in these women, the body mass index was in the range from 30 to 40 kg/m2).
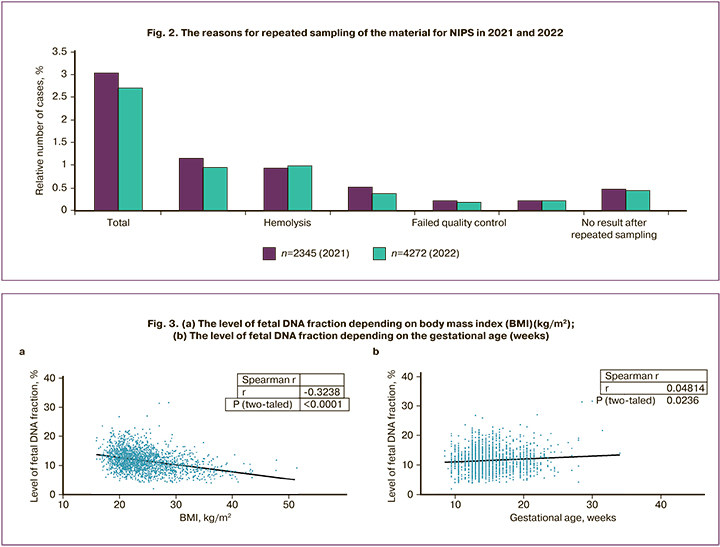
According to literature data, low fetal DNA content is more often observed in overweight and obese women, and is also associated with pregnancy [13, 15, 20]. A similar trend was observed in our group; at the same time, we did not detect a remarkable dependence in the content of FF on the gestational age. We carried out a correlation analysis of the dependence of the FF level on the body mass index and on the gestational age. Since the distribution of parameters in both cases is not normal, Spearman’s nonparametric criterion was used to determine the correlation. The strength of the correlation coefficient of the FF level with the body mass index is moderate negative (-0.324) with a high level of significance (p<0.0001) (Fig. 3a). The strength of the correlation coefficient of the FF level with the gestational age is very weak (0.048) with a significance level of 0.236 (Fig. 3b).
Frequency of chromosomal abnormalities detected by NIPS
Representation of chromosomal abnormalities
The sample of patients examined in this study is not population sample. In this regard, the data obtained on its basis do not allow us to assess completely the effectiveness of NIPS in comparison with the first trimester prenatal screening; however, the division of this sample into subgroups according to certain indications and the study of the spectrum of detected abnormalities is of some value.
According to the indications for NIPS, we divided all pregnant women into four groups:
- a high-risk group of fetal chromosomal pathology according to the results of the first trimester prenatal screening (≥1:100) 774/4272 (18.12%);
- a group of women with an intermediate risk of fetal chromosomal pathology according to the results of the first trimester prenatal screening (1:101–1:1000) 1666/4272 (39.00%);
- women who are concerned about the condition of the fetus, whose risk of aneuploidy is lower than the population according to the results of the first trimester prenatal screening (≤1:1000) 1532/4272 (35.86%);
- pregnant women who had a risk of fetal chromosomal abnormality in the second trimester, whereas the risk was not detected in the first trimester, or there was no screening of the first trimester 300/4272 (7.02%).
Table 2 shows the data on the groups of pregnant women who underwent a non-invasive prenatal study.
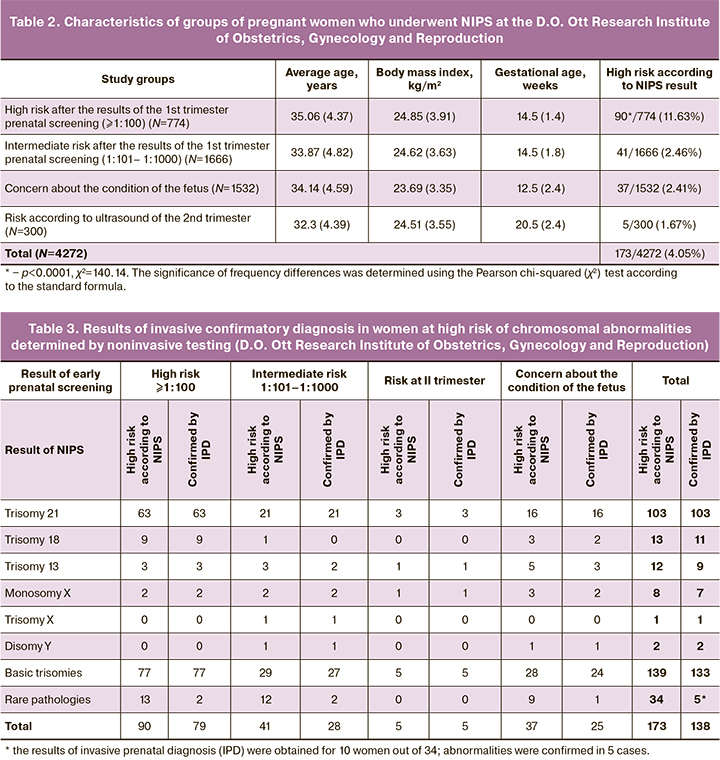
Most of the pregnant women who had NIPS demonstrated the so–called intermediate risk according to the results of the first trimester prenatal screening, namely, 1666/4272 (39.00%), and these results are consistent with the mission of this test. The group of women concerned about the condition of the fetus, 1532/4272 (35.86%), were the second in this rating.
The age and body mass index of pregnant women in different groups are similar in parameters, but the gestational age of the patients is different. The highest gestational age was 20.5 weeks in the group of women at risk of the second trimester. The average gestational age did not differ significantly in the groups of women at high and intermediate risk (the mean value was 14.74 and 15.11 weeks, respectively). The average gestational age in the group of women concerned about the condition of the fetus was 12.5 weeks and it was lower than in all the above-mentioned groups.
In the group of women with a high risk of chromosomal pathology, according to the results of first trimester prenatal screening, the number of detected pathologies was 90/774 (11.63%), which statistically significantly higher than the frequency of aneuploidy compared to the total number of other groups p<0.0001, χ2=140.14. There were 37/1532 (2.41%) and 41/1666 (2.46%) samples among pregnant women concerned about the condition of the fetus and women with an intermediate risk, respectively. Aneuploidy was detected in only five cases, 5/300 (1.67%), in the group of women at increased risk in the second trimester.
It should be particularly noted that gestational age in group of women concerned about the condition of the fetus was up to 12 weeks and 6 days in 743/1552 (47.87%) cases. Chromosomal pathologies were detected in 24 of them, which are approximately 2/3 of the total number of detected aneuploidies in this group (37).
Effectiveness of NIPS for detecting aneuploidies
We identified 173 aneuploidies in this study including 139 which are the most common chromosomal abnormalities (aneuploidies in chromosomes 21, 18, 13, X and Y compatible with postnatal development); these resulted accounted for 80.3% of all identified variants (Table 3). However, if we count the number of confirmed pathologies, then the proportion of major chromosomal abnormalities increased to 96.4% (133/138). Such a large gap in numbers may be associated with the presence of placental mosaicism. All pathologies identified by us were analyzed twice in order to exclude a possible error. The data were reproduced in 100% of cases and are presented in Table 3.
The results of NIPS were confirmed by invasive diagnosis (amniocentesis) for all 103 fetuses with Down syndrome (thus, the probability of trisomy 21 with a positive test result is 100%, false negative result (FNR) = 0%, false positive result (FPR) = 0%). After invasive diagnostics, two FPR were identified out of 13 cases with a risk of trisomy 18 (positive predictive value of the test was 84.6%, FNR=0%, FPR=15.4%). Three more FPR were detected by an invasive method out of 12 cases with a risk of trisomy 13 (positive predictive value of the test was 75%, FNR=0%, FPR=25%). One FPR was revealed among 11 fetuses with sex chromosome abnormalities (positive predictive value of the test was 90.9%, FNR=0%, FPR=9.1%). Thus, it is worth noting that our study identified a lower number of FPR in sex chromosomes in comparison with the data of the studies published by the Russian researchers.
As a result, we revealed the following abnormalities after invasive confirmatory diagnosis: trisomy 21 (Down syndrome) in 103/138 cases (74.63%), trisomy 18 (Edwards syndrome) in 11/138 cases (7.97%), trisomy 13 (Patau syndrome) in 9/138 cases (6.52%). The analysis of sex chromosomes detected the risk of trisomy X in one case, monosomy X in eight (5.79%) cases and disomy Y in two male fetuses (Jacobs syndrome).
The highest percentage of FPR for all aneuploidies was in the group of patients concerned about the condition of the fetus (14.8%) followed by the intermediate risk group (6.9%). FPR for common chromosomal abnormalities were not revealed in the high-risk group and in the risk group in the second trimester.
Noninvasive testing also detected a high risk of a rare pathology: trisomy 2, 3, 5, 11, 12, 14, 15 (one case for each pathology), trisomy 8, 9, 20, 10, 22 (in two cases), trisomy 19 (in three cases), trisomy 16 (ten cases); monosomy 21, 18 and 13, as well as the risk of multiple aneuploidies in one case. In our study, most aneuploidies were detected in chromosomes 16 and 19. Trisomy 16 in the placenta may be associated with placental insufficiency, fetal growth retardation. Trisomy 19 can be detected in patients with malignant neoplasms of the blood. Isolated cases of detecting trisomy 19 during pregnancy are described in the literature.
Since fetal DNA in the mother’s bloodstream is a product of the placenta, there is limited placental mosaicism in most cases, which can be considered as a risk factor for pregnancy complications. In all these cases, the decision on invasive diagnostis was made by the pregnant women after consulting the doctor and analyzing the results of ultrasound assessment. Most women with rare aneuploidies refused to have invasive diagnostics if there were no ultrasound markers of chromosomal abnormalities.
After the ultrasound assessment revealed congenital malformations of the fetus in a pregnant woman with a risk of trisomy 2, the pregnancy was terminated and the chromosomal pathology in the fetus was confirmed by karyotyping. Trisomy 22 was confirmed during the study of the material of missed miscarriage. When monosomy 18 was suspected, karyotyping established a deletion of the long arm of chromosome 18 in the fetus. Only two out of ten women with suspected trisomy 16 agreed to have invasive diagnosis; trisomy 16 was detected in the fetus in one case. Monosomy 21 in the fetus was not confirmed; apparently, placental mosaicism was observed in the latter cases.
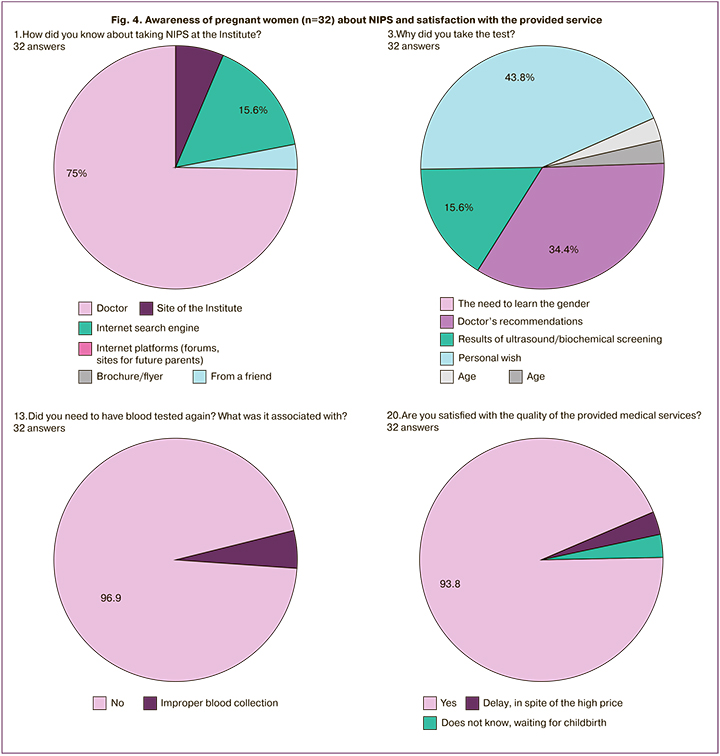
Awareness of pregnant women about NIPS and satisfaction with the provided service
An important parameter in assessing the quality of any study is the attitude of patients. In order to find out their attitude, we conducted a small survey. There were several questions and the most significant of them were the following:
- How did you know about the test?
- Why did you take the test?
- Did you need to have blood tested again; did the doctors explain it to you?
- Are you satisfied with the service and will you recommend the Institute to colleagues and friends?
The answers to these questions are shown in Figure 4. The survey of the patients’ opinion showed that they learned about the possibility of having NIPS primarily from their doctor, then from the internet. Less frequently, pregnant women found information on the Institute’s website or learned from friends. Patients did not pay attention to the purpose of the test in 43.8% of cases, that means they could choose the test not according to its function, but due to its price. This screening was administered largely on the basis of personal initiative (43.8% compared to 34.4% of the doctor’s opinion). The test was performed for the first time in 97% of cases. Blood had to be taken again in 3% of cases due to the lack of fetal biological material in the primary sample, however, the patients were not fully informed about this. Most patients (more than 93%) were satisfied and are ready to recommend the Institute to their colleagues. Less than 3% were not satisfied with the timing of the study or they are waiting for childbirth to make their choice.
Discussion
At the present stage of development of molecular genetic technologies, high throughput sequencing of extracellular DNA of a pregnant woman is widely used for conducting NIPS. Nowadays, this method is becoming part of routine clinical practice which makes it possible to detect in the fetus not only common trisomy 13, 18, 21, aneuploidy in sex chromosomes, but also partial trisomy, deletions, placental mosaicism. This method also has prospects as a method for assessing the risk of certain pathologies in the mother [13, 21].
In our study we again confirmed that NIPS is appropriate for almost all categories of pregnant women at any gestation period starting from 8.5 weeks.
The advanced average age of pregnant women who took part in this study (35 years; 60% of all participants are women from 30 to 40 years old) indicates a conscious choice of this technology (both by the patient and by the doctor). It is evident that this test is necessary primarily for patients of the older age group, where the risk of chromosomal pathology increases. This suggestion is also confirmed by the results of a post-test survey of satisfaction with NIPS where personal initiative was the leading motivation for conducting the screening.
It is quite obvious that this test was done at 12–15 weeks gestation as it corresponds to the logic of its administration. Only in some cases (121/4272 (2.82%)) it was carried out at 22 weeks gestation or later. The administration of this test at a later period was associated with missed early prenatal screening and the woman’s personal initiative in most cases.
It also should be noted that we observed a special group of patients: more than a third of women, namely 1495/4272 (35.0%), were obese or overweight. It is important that some of them (26 women) failed to have NIPS during the initial examination. After repeated blood sampling, only four women did not have the results of the test. Thus, despite the fact that there is a direct negative relationship between the success of the test and the body mass index [13], our technology allows obese or overweight women to solve their screening tasks effectively in 99.7% of cases.
A total of 19 women out of 4272 did not receive NIPS results (0.44% or even 0.33% if we exclude women who refused to do the test again from the study sample); this parameter is the lowest among the findings of the previous Russian studies [17, 18]. Despite the link between the level of our laboratory and decrease in the number of cases which required repeated sampling for NIPS, such a low percentage of the absence of results cannot be explained only by the increased experience of our laboratory (other Russian laboratories had a much larger number of tests). Probably, the details are associated with the technology itself, initial validation of the algorithm on control samples which were collected at the Institute and tested in other centers, a unique bioinformatic algorithm and attentive attitude to patients (94% of patients are completely satisfied with the test).
The analysis of repeated sampling of the material for NIPS indicates that determining the gender of the fetus is the most challenging task for the test and it is even more significant than the problem of a low FF level. It is possible to obtain the required percentage of FF (more than 4%) with repeated blood sampling in 90% of cases, while the gender of the fetus can be precisely determined in less than half of cases. Thus, the main problem of the test remains identification of the gender of the fetus, according to the literature data [22, 23] and our own experience; this difficulty leads to the inability to make a conclusion, and/or a limitation for the accurate detection of abnormalities associated with sex chromosomes.
This limitation of NIPS may result in the refusal of a number of researchers and clinicians from the analysis of chromosomal abnormalities of the gender and focus on screening only three of the most common pathologies: trisomy 21, 13 and 18 [24]. However, the sensitivity of the test (its ability to detect fetal pathology) decreases in this case, and noninvasive screening conducted with the help of this test can no longer be considered as an alternative to early trimester prenatal screening. The results of our study also demonstrate this dilemma. In particular, we have shown that the detected abnormalities on trisomy 21, 18 and 13 together account for not more than 75% (128/173) of the total pathology, which is not enough. The largest percentage among them is Down syndrome, namely 103/173 (60%), which is consistent with the results of the Russian and international studies [4, 5, 25]. About 21% (36/173) of abnormalities are trisomy 13, 18 and sex chromosomes, and 19% (34/173) are rare trisomies [26, 27]. In terms of the number of confirmed cases, trisomy 21 (Down syndrome) remains the most common chromosomal pathology, its share increases to 75% (103/138); while the share of trisomy 18 (Edwards syndrome), trisomy 13 (Patau syndrome) and disorders in sex chromosomes remains at the same level, 21% (30/138) cases. Such changes are explained by the fact that all cases of trisomy 21 in our study were confirmed by invasive prenatal diagnosis, whereas other common trisomies were confirmed in less than 77.5% of cases (the number of FPR ranged from 12.5 to 25%). Moreover, the largest number of FPR was noted in the study of trisomy 13 and 18, and not sexual chromosomes, and this contradicts the global data [3]. It is possible that the low number of FPR on sex chromosomes is associated with a small sample and an increase in their number should be expected in the future.
The analysis of all FPR showed that the largest number of them was noted in the group of those concerned about the condition of the fetus (14.8%) followed by the group of patients at intermediate risk (6.9%). In the high-risk group and in the risk group of women at the 2nd trimester, FPR on the most frequent chromosomal abnormalities was not detected. This distribution seems to be quite reasonable and is due to the heterogeneity of the first two groups.
The group of patients concerned about the condition of the fetus is an interesting sample of patients. They do not differ in age or body mass index from other groups of patients. However, they are different in average gestational age. Most pregnant women decided to participate in testing at 12.5 weeks gestation. And it also should be noted that 2/3 (24/37) of the total number of detected aneuploidies in this group were revealed in women who had the test before 12 weeks. This fact is difficult to explain, since only in 3 out of 24 cases there was an early miscarriage associated with reproductive losses that could statistically not reach other groups. In any case, a large number of abnormalities in this group, especially those detected at an early stage, is a definite challenge to the proposed contingent model [8], since this group of pregnant women will not be taken into account when using such a model. At this stage, it is also impossible to develop objective criteria that could be used for screening pregnant women who are concerned about the condition of the fetus free of charge that is at the expense of the state. However, pregnant women who paid for the test by themselves and who received a positive test (only them) can be integrated into the new screening algorithm as follows: such patients do not need to undergo biochemical screening, but need ultrasound and subsequent referral for invasive prenatal diagnosis.
Conclusion
The largest number of the detected chromosomal abnormalities in fetal development was found in the high-risk group of patients. The proportion of such pregnant women in our study is insignificant. Screening in this group was performed mainly due to the patient’s concerns about invasive interventions. After introducing the contingent model, the number of such patients is likely to decrease. The comparable number of chromosomal abnormalities in groups of patients concerned about the condition of the fetus and intermediate risk emphasizes not only the importance of screening of pregnant women at intermediate risk, but also the need to take into account the screening programs of pregnant women who are concerned about the condition of the fetus.
The necessity of using a genome-wide NIPS remains controversial. On the one hand, a rare pathology associated with other chromosomal abnormalities is confirmed in 50% of cases (according to our data), however, the majority of patients with such findings refuse further invasive prenatal diagnosis and complicate the analysis of the appropriateness of detecting these chromosomal abnormalities.
NIPS has been frequently used in practice. In spite of the high price, many women want to take it, but at the same time they rely on their opinion to a greater extent than on the opinion of a doctor (an obstetrician-gynecologist, not a geneticist, has a much greater influence on them).
It is worth noting that NIPS which was performed using the bioinformatic analysis is a reliable method that makes it possible to determine precisely the chromosomal pathologies of the fetus, both regular trisomies, aneuploidies in the sex chromosomes, and “random findings”. And given the expected further reduction in the cost of this type of research, we assume that the NIPS of fetal aneuploidy using genome-wide sequencing will become more common and will be included in new algorithms for pregnancy screening.
References
- Баранов В.С., Кузнецова Т.В., Кащеева Т.К., Иващенко Т.Э. Пренатальная диагностика наследственных болезней. Состояние и перспективы. Санкт-Петербург: Эко-Вектор; 2020. 469с. [Baranov V.S., Kuznetsova T.V., Kashcheyeva T.K., Ivashchenko T.E. Prenatal diagnosis of hereditary diseases. State and prospects. St. Petersburg: Eko-Vector Publishing House; 2020. 469p. (in Russian)].
- Hook E.B. Chromosome abnormalities: prevalence, risks and recurrence. In: Brock D.L.H., Rodeck C.H., Ferguson-Smith M.A., eds. Prenatal diagnosis and screening. Edinburgh: Churchill Livingstone; 1992: 351-92.
- Parker M.J., Budd J.L.S., Draper E.S., Young I.D. Trisomy 13 and trisomy 18 in a defined population: epidemiological, genetic and prenatal observations. Prenat. Diagn. 2003; 23(10): 856-60. https://dx.doi.org/10.1002/pd.707.
- Nicolaides K.H. Screening for fetal aneuploidies at 11 to 13 weeks. Prenat. Diagn. 2011; 31(1): 7-15. https://dx.doi.org/10.1002/pd.2637.
- Баранов В.С., Кузнецова Т.В. Цитогенетика эмбрионального развития человека. Научно-практические аспекты. СПб: Н-Л; 2007. 640с. [Baranov V.S.,Kuznetsova T.V. Cytogenetics of human embryonic development. Scientific and practical aspects. St. Petersburg: N-L Publisher; 2007. 640p.(in Russian)].
- Айламазян Э.К., Баранов В.С., ред. Пренатальная диагностика наследственных и врожденных болезней. СПб.: МЕДпресс-информ; 2006. 415c. [Ailamazyan E.K., Baranov V.S., ed. Prenatal diagnosis of hereditary and congenital diseases. St. Petersburg: MEDpress-inform; 2006. 415 p.(in Russian)].
- Сухих Г.Т., Трофимов Д.Ю., Барков И.Ю., Донников А.Е., Шубина Е.С., Коростин Д.О., Екимов А.Н., Гольцов А.Ю., Бахарев В.А., Каретникова Н.А., Боровиков П.И., Тетруашвили Н.К., Ким Л.В., Гата А.С., Павлович С.В., Скрябин К.Г., Прохорчук Е.Б., Мазур А.М., Пантюх К.С. Неинвазивный пренатальный ДНК-скрининг анеуплоидий плода по крови матери методом высокопроизводительного секвенирования. Клинические рекомендации. Акушерство и гинекология. 2016; 6: 129-57. https://dx.doi.org/10.18565/aig.2016.6.recomendations. [Sukhikh G.T., Trofimov D.Yu.,Barkov I.Yu. et al. Noninvasive prenatal DNA screening of fetal aneuploidies from maternal blood by high-throughput sequencing. Clinical guidelines. Obstetrics and Gynecology. 2016; 6: 129-57. (in Russian)]. https://dx.doi.org/10.18565/aig.2016.6.recomendations.
- Калашникова Е.А., Глотов А.С., Андреева Е.Н., Барков И.Ю., БобровникГ.Ю., Дубровина Е.В., Жученко Л.А. Современное значение неинвазивного пренатального исследования внеклеточной ДНК плода в крови материи перспективы его применения в системе массового скрининга беременных в Российской Федерации. Журнал акушерства и женских болезней. 2021; 70(1): 19-50. https://dx.doi.org/10.17816/JOWD56573.[Kalashnikova E.A., Glotov A.S., Andreyeva E.N., Barkov I.Yu.,Bobrovnik G.Yu., Dubrovina E.V., Zhuchenko L.A. Current relevance of non-invasive prenatal study of cell-free fetal DNA in the mother’s blood and prospects for its application in mass screening of pregnant women in the Russian Federation. Journal of Obstetrics and Women’s Diseases. 2021;70(1):19-50.(in Russian)]. https://dx.doi.org/10.17816/JOWD56573.
- Sparks A.B., Struble C.A., Wang E.T., Song K., Oliphant A. Non-invasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: evaluation for trisomy 21 and trisomy 18. Am. J. Obstet. Gynecol. 2012; 206(4): 319.e1-9. https://dx.doi.org/10.1016/j.ajog.2012.01.030.
- Fan H.C., Blumenfeld Y.J., Chitkara U., Hudgins L., Quake S.R. Non-invasive diagnosis fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA. 2008; 105(42): 16266-71. https://dx.doi.org/10.1073/pnas.0808319105.
- Bianchi D.W., Parker RL., Wentworth J., Madankumar R., Saffer C., Das A.F. et. al. DNA sequencing versus standard prenatal aneuploidy screening. N. Engl. J. Med. 2014; 370(9): 799-808. https://dx.doi.org/10.1056/NEJMoa1311037.
- Ericsson O., Ahola T., Dahl F., Karlsson F., Persson F., Karlberg O. et al. Clinical validation of a novel automated cell-free DNA screening assay for trisomies 21, 13, and 18 in maternal plasma. Prenat. Diagn. 2019; 39(11): 1011-5.https://dx.doi.org/10.1002/pd.5528.
- Кудрявцева Е.В., Ковалёв В.В., Баранов И.И., Канивец И.В., Киевская Ю.К., Коростелёв С.А. Низкая фетальная фракция внеклеточной ДНК при проведении неинвазивного пренатального ДНК-скрининга: возможные причины, клиническое значение и тактические решения. Доктор.Ру.2020; 19(8): 49-54. https://dx.doi.org/10.31550/1727-2378-2020-19-8-49-54. [Kudryavtseva E.V., Kovalev V.V., Baranov I.I., Kanivets I.V., Kievskaya Yu.K., Korostelev S.A. Low Fetal Fraction of Cell-free DNA Identified by Non-invasive Prenatal DNA Testing: Possible Causes, Clinical Significance, and Tactics. Doctor.Ru. 2020; 19(8): 49-54. (in Russian)].https://dx.doi.org/10.31550/1727-2378-2020-19-8-49-54.
- Taglauer E.S., Wilkins-Haug L., Bianchi D.W. Review: cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta. 2014; 35(Suppl.): S64-8. https://dx.doi.org/10.1016/j.placenta.2013.11.014.
- Qiao L., Zhang Q., Liang Y., Gao A., Ding Y., Zhao N. et al. Sequencing of short cfDNA fragments in NIPT improves fetal fraction with higher maternal BMI and early gestational age. Am. J. Transl. Res. 2019; 11(7): 4450-9.
- Ковалева Ю.А., Хасанов А.А., Сингатуллина Л.М. Определение внеклеточных ДНК крови – клиническое и диагностическое значение. Практическая медицина. 2010; 4: 63-6. [Kovaleva Y.A., Khasanov A.A., Signatullina L.M. Determination of extracellular DNA in the blood – clinical and diagnostic value. Practical medicine. 2010; 4: 63-6.(in Russian)].
- Оленев А.С., Баранова Е.Е., Беленикин М.С., Галактионова А.М., Гнетецкая В.А., Зобкова Г.Ю., Кузнецова Е.С., Макарова М.В., Сагайдак О.В., Сонголова Е.Н. Внедрение неинвазивного пренатального теста (НИПТ) в структуру пренатальной диагностики г. Москвы. В кн.: VII Всероссийская конференция с международным участием «Геномная медицина в пренатальной диагностике, генетическом паспорте и генной терапии». Санкт-Петербург, 12-13 ноября 2020г. СПб.; 2020: 99-103. [Olenev A.S., Baranova E.E.,Belenikin M.S., Galaktionova A.M., Gnetetskaya V.A., Zubkova G.Yu. et al. Introduction of noninvasive prenatal test (NIPT) into the structure of prenatal diagnostics in Moscow. Genomic medicine in prenatal diagnostics, genetic passport and gene therapy. Saint Petersburg; 2020: 99-103. (in Russian)].
- Кудрявцева Е.В., Канивец И.В., Киевская Ю.К., Баранов И.И., Ковалев В.В., Коростелев С.А. Неинвазивный пренатальный тест в России: популяционное исследование. Акушерство и гинекология. 2019; 12: 30-5.https://dx.doi.org/10.18565/aig.2019.12.30-35. [Kudryavtseva E.V., Kanivets I.V., Kievskaya Yu.K., Baranov I.I., Kovalev V.V., Korostelev S.A. Noninvasive prenatal testing in Russia: a population study. Obstetrics and Gynecology. 2019; 12: 30-35. (in Russian)]. https://dx.doi.org/10.18565/aig.2019.12.30-35.
- Wright D., Wright A., Nicolaides K.H. A unified approach to risk assessment for fetal aneuploidies. Ultrasound Obstet. Gynecol. 2015; 45(1): 48-54.https://dx.doi.org/10.1002/uog.14694.
- Committee Opinion No. 640: Cell-free DNA screening for fetal aneuploidy. Obstet. Gynecol. 2015; 126(3): e31. https://dx.doi.org/10.1097/AOG.0000000000001051.
- Сухих Г.Т., Тетруашвили Н.К., Трофимов Д.Ю., Ким Л.В., Барков И.Ю.,Шубина Е.C., Парсаданян Н.Г., Федорова Н.И., Гольцов А.Ю., Александрова Н.В. Неинвазивный пренатальный ДНКскрининг методом высокопроизводительного секвенирования у беременных с акушерской патологией. Доктор.Ру. 2017; 3: 11-5. [Sukhikh G.T., Tetruashvili N.K., Trofimov D.Yu., Kim L.V., Barkov I.Yu., Shubina Ye.S., Parsadanyan N.G., Fedorova N.I., Goltsov A.Yu., Alexandrova N.V. Next-Generation Sequencing Technologies As a Noninvasive Prenatal DNA Screening Method in Pregnant Women with Obsteric Disorders. Doctor.Ru. 2017; 3(132): 11-15. (in Russian)].
- Soukkhaphone B., Lindsay C., Langlois S., Little J., Rousseau F., Reinharz D. Non-invasive prenatal testing for the prenatal screening of sex chromosome aneuploidies: A systematic review and meta-analysis of diagnostic test accuracy studies. Mol. Genet. Genomic Med. 2021; 9(5): e1654. https://dx.doi.org/10.1002/mgg3.1654.
- Zhang B., Lu B.Y., Yu B., Zheng F.X., Zhou Q., Chen Y.P., Zhang X.Q. Noninvasive prenatal screening for fetal common sex chromosome aneuploidies from maternal blood. J. Int. Med. Res. 2017; 45(2): 621-30.https://dx.doi.org/10.1177/0300060517695008.
- Hörmansdörfer C., Schmidt P., Hillemanns P., Scharf A. Die pränatale Detektion der Trisomien 13, 18 und 21: Vergleich des Advanced First Trimester Screenings (AFS) mit dem Ersttrimester-Screening nach Nicolaides [The prenatal detection of trisomy 13, 18, and 21: comparison of the advanced first trimester screening (AFS) with the first trimester screening according to Nicolaides]. Z. Geburtshilfe Neonatol. 2007; 211(6): 243-9. (in German). https://dx.doi.org/10.1055/s-2007-981361.
- Кудрявцева Е.В., Ковалёв В.В., Канивец И.В., Киевская Ю.К., Коростелёв С.А. Free-DNA плода: опыт популяционного скрининга хромосомной патологии в России. Вопросы гинекологии, акушерства и перинатологии. 2019; 18(3): 46-51. [Kudryavtseva E.V., Kovalev V.V., Kanivets I.V., Kievskaya Yu.K., Korostelev S.A. Cell-free-fetal DNA: an experience of population screening for chromosome pathology in Russia. Issues of Gynecology, Obstetrics and Perinatology. 2019; 18(3): 46-51. (in Russian)].
- Scott F., Bonifacio M., Sandow R., Ellis K., Smet M.E., McLennan A. Rare autosomal trisomies: important and not so rare. Prenat. Diagn. 2018; 38(10): 765-71. https://dx.doi.org/10.1002/pd.5325.
- Оленев А.С., Баранова Е.Е., Сагайдак О.В., Кузнецова Е.С., Галактионова А.М., Капланова М.Т., Беленикин М.С., Гнетецкая В.А., Сонголова Е.Н. Случайные находки при использовании полногеномного неинвазивного пренатального теста: клинические и этические аспекты. Проблемы репродукции. 2021; 27(1): 78 87. https://dx.doi.org/10.17116/repro20212701178. [Olenev A.S., Baranova E.E., Sagaydak O.V.,Kuznetsova E.S., Galaktionova A.M., Kaplanova M.T., Belenikin M.S., Gnetetskaya V.A., Songolova E.N. Random findings in the use of a whole genome noninvasive prenatal test: clinical and ethical aspects. Russian Journal of Human Reproduction. 2021;27(1):78 87. (in Russian)]. https://dx.doi.org/10.17116/repro20212701178.
Received 17.06.2022
Accepted 17.10.2022
About the Authors
Olga A. Tarasenko, Ph.D. (Bio), Researcher at the Laboratory of Genomics, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology, +7(921)923-91-41, olgatar777@gmail.com, https://orcid.org/0000-0003-3394-7391, 199034, Mendeleevskaya line, 3, St. Petersburg, Russia.Elena S. Vashukova, Ph.D. (Bio), Researcher at the Laboratory of Genomics, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology,
vi_lena@list.ru, https://orcid.org/0000-0002-6996-8891, 199034, Mendeleevskaya line, 3, St. Petersburg, Russia.
Polina Yu. Kozyulina, bioinformatician, “NIPT”, polykoz@gmail.com, 199178, emb. Reki Smolenki, 5-7, St. Petersburg, Russia.
Alisa V. Morshneva, bioinformatician, “NIPT”, 1195alisa@gmail.com, 199178, emb. Reki Smolenki, 5-7, St. Petersburg, Russia.
Anastasia R. Maltseva, Research Assistant at the Laboratory of Genomics, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology,
nastya.chentsova@gmail.com, 199034, Mendeleevskaya line, 3, St. Petersburg, Russia.
Olga V. Pachulia, Ph.D., Scientific Secretary, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology, for.olga.kosyakova@gmail.com,
199034, Mendeleevskaya line, 3, St. Petersburg, Russia.
Olga E. Talantova, Ph.D., Senior Researcher at the Laboratory of Genomics, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology,
olga_talantova@mail.ru, https://orcid.org/0000-0003-3520-599X, 199034, Mendeleevskaya line, 3, St. Petersburg, Russia.
Alexander L. Koroteev, Ph.D., Head Physician, Diagnostic Center (Medical and Genetic), alexkoroteev@mail.ru, 194044, Tobolskaya str., 5, St. Petersburg, Russia.
Olesya N. Bespalova, Dr. Med. Sci., Deputy Director for Research, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology, shiggerra@mail.ru,
199034, Mendeleevskaya line, 3, St. Petersburg, Russia.
Igor Yu. Kogan, Dr. Med. Sci., Head of the D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology, ikogan@mail.ru,
https://orcid.org/0000-0002-7351-6900, 199034, Mendeleevskaya line, 3, St. Petersburg, Russia.
Andrey S. Glotov, Dr. Bio. Sci., Head of the Department of Genomic Medicine, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology, anglotov@mail.ru, https://orcid.org/0000-0002-7465-4504, 199034, Mendeleevskaya line, 3, St. Petersburg, Russia.
Corresponding author: Olga A. Tarasenko, olgatar777@gmail.com
Authors’ contributions: Tarasenko O.A., Vashukova E.S., Glotov A.S. – developing the concept and design of the study;
Tarasenko O.A., Talantova O.E., Maltseva A.R., Vashukova E.S., Pachulia O.V., Ivashchenko T.E. – collecting and processing the material; Kozyulina P.Yu., Morshneva A.V. – bioinformatic processing of the results; Tarasenko O.A., Vashukova E.S. – statistical data processing; Tarasenko O.A., Vashukova E.S., Glotov A.S., Baranov V.S. – text writing; Tarasenko O.A.,
Vashukova E.S., Koroteev A.L., Bespalova O.N., Kogan I.Yu., Baranov V.S., Glotov A.S. – editing the text of the article.
Conflicts of interest: The authors declare that there are no conflicts of interest.
Funding: Processing of the results of the study was carried out with the support of the scientific program No. AAAAA-A20-120041390028-0.
Ethical Approval: The study was approved by the Ethical Review Board of the D.O. Ott Research Institute of Obstetrics, Gynecology and Reproduction, St. Petersburg, Russia (Protocol No.130 – 16/07/2020).
Patient Consent for Publication: All patients participating in the study provided an informed consent for the publication of their data.
Authors’ Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator.
For citation: Tarasenko O.A., Vashukova E.S., Kozyulina P.Yu., Morshneva A.V., Maltseva A.R., Pachulia O.V., Talantova O.E., Koroteev A.L., Ivashchenko T.E., Bespalova O.N., Kogan I.Yu., Baranov V.S., Glotov A.S. Experience of using high-throughput sequencing (NGS) for noninvasive prenatal screening of fetal aneuploidy at the D.O. Ott
Research Institute of Obstetrics, Gynecology and Reproduction.
Akusherstvo i Ginekologiya /Obstetrics and Gynecology. 2022; 10: 37-49 (in Russian)
https://dx.doi.org/10.18565/aig.2022.10.37-49



