Embryo quality assessment by the small noncoding RNA expression profile in an embryo culture medium in assisted reproductive technology programs
Objective. To investigate of the small noncoding RNA (sncRNA) (microRNA and piwiRNA) profile in an embryo culture medium on day 4 of cultivation to identify the potential biomarkers for embryo quality.Timofeeva A.V., Kalinina E.A., Drapkina Yu.S., Chagovets V.V., Makarova N.P., Sukhikh G.T.
Materials and methods. The deep sequencing method was used to identify the spectrum of sncRNAs present in the blastocyst cavity and embryo culture medium, which were quantified using a real-time reverse-transcription polymerase chain reaction assay in 48 samples of the culture medium of embryos transferred into the uterine cavity.
Results. Partial least squares-discriminant analysis (PLS-DA) was used to analyze the contribution of six sncRNAs to determining the embryo quality, which was characterized by the variable importance in projection (VIP) values. The analysis showed that the greatest contribution was made by four sncRNAs: piR020401 (VIP = 1.46027), let-7c-5p (VIP = 1.17416), let-7b-5p (VIP = 0.994657), and let-7i-5p (VIP = 0.942665), and the smallest contribution was by miR-143-5p (VIP = 0.623108) and miR-92a-3p (VIP = 0.471951). The Spearman rank correlation method was used to reveal a statistically significant correlation between the expression level of sncRNA (hsa-let-7b-5p, hsa-let-7c-5p, and hsa-piR020401) in the culture medium and the quality of the embryo transferred to the uterine cavity. The Mann-Whitney non-parametric test revealed statistically significant differences in the expression level of piR020401 and miR-92a-3p in the culture medium of embryos forming groups according to morphofunctional parameters.
Conclusion. The sncRNAs (let-7c-5p, let-7b-5p, piR020401) found in the culture medium statistically significantly correlate with embryo morphofunctional characteristics on day 4 after fertilization, among which piR020401 makes the greatest contribution to determining the embryo quality. The expression level of miR-92a-3p and piR020401 in the culture medium depends on the stage of embryonic development, differentiating the morula and blastocyst stage. Considering that the lag in the embryonic development rate by day 5 of cultivation did not always predetermine the absence of embryo implantation into the uterus, it is not advisable to focus exclusively on embryo morphological parameters when choosing a candidate for transfer to the uterus, and it is recommended that the expression profile of let-7c-5p, let -7b-5p, miR-92a-3p, and piR020401 should be additionally estimated on day 4 of embryo cultivation. Thus, the findings make it possible to optimize the choice of an embryo and to implement an assisted reproductive technology program.
Keywords
Nowadays, a very high level of infertility leads to the rapid development of assisted reproductive technologies (ART) [1]. Despite of many successful attempts aimed at demographic situation improvement, the percentage of infertile couples in Russia ranges between 8 to 17.5% and is not currently a downward trend [2]. Today ART plays the leading role in the treatment of different types of infertility.
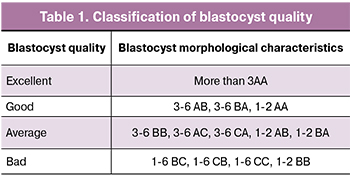 To achieve pregnancy in IVF program the most challenging task is the embryo selection. The embryo should be of the highest quality and viability. Selection of such embryos is usually carried out according to their morphological properties evaluation. The generally accepted standard for assessing blastocysts quality was proposed by Gardner et al. [3] and is demonstrated in Table 1.
To achieve pregnancy in IVF program the most challenging task is the embryo selection. The embryo should be of the highest quality and viability. Selection of such embryos is usually carried out according to their morphological properties evaluation. The generally accepted standard for assessing blastocysts quality was proposed by Gardner et al. [3] and is demonstrated in Table 1.
However, Gardner et al. grading classification is subjective and the accuracy of this embryo selection method is not high enough. Moreover, not all embryos of good and excellent quality, when transferred into the normal endometrium, result in pregnancy [4]. Thus, there has been a need for introduction of additional non-invasive methods of selection of the embryo with the highest implantation potential.
In recent years a significant number of potential biomarkers of embryo quality and implantation potential have been proposed. A promising method is preimplantation genetic testing, since chromosomal abnormality in transferred embryos can be a major cause of negative results. However, along with the undisputed value, this method has a limited application in routine clinical practice due to its high cost, as well as the requirement of invasive interventions on the embryo [5].
The study of energy, metabolic activity and signalling pathways of a particular embryo without any invasive procedures is possible with the help of embryo culture medium obtained at various embryo development stages [6]. Small noncoding RNAs (sncRNAs) have a significant diagnostic and prognostic potential in relation to the embryo quality and implantation ability. These molecules can be detected in embryo culture medium [7]. SncRNAs act on the epigenetic, transcriptional and post-transcriptional levels of the gene regulation expression. These interactions determine the different cell phenotype and functions [8]. SncRNAs like microRNA and piwiRNA have the significant influence on cell phenotype and functions determination [9]. It has been shown that the detection of these molecules in the embryo culture medium makes it possible to predict embryo implantation potential and to assess the quality of this embryo more accurately [10]. In 2014, McCallie et al. [11] were the first to show that blastocysts in patients with PCOS as an infertility factor have atypical microRNA profile (miR-19a, miR-19b, miR-24, miR-92, miR-93, etc.), and in 2016, Capalbo et al. [12] demonstrated that intracellular microRNA expression differs in euploid and aneuploid embryos. In the study of Liang et al. [13], it was proven that active expression of miR-24 in embryo culture medium was associated with the blastocyst development failure and had an impact on embryo quality. Thus, a relationship between the quality of embryo and specific sncRNA profile in embryo culture medium is very promising and innovative as a method of embryo selection.
Materials and Methods
There were 49 samples of spent embryo culture medium which were obtained on day 4 after fertilisation during IVF cycles from embryos A, B and C quality, according to Gardner grading scale and also from morulas and cavernous morulas. Morphological characteristics of studied embryos were evaluated and were as follows: blastocysts of excellent (А, n = 27), good (B, n = 8) and bad (C, n = 4) quality. The remaining 10 samples of culture media were obtained from 7 morulas and 3 cavernous morulas. RNAs were isolated from the collected samples by the column method using the miRNeasy Serum/Plasma Kit (Qiagen). Identification of all sncRNAs in embryo culture media was carried out with deep sequencing analysis using cDNA Library NEBNext® Multiplex Small RNA Library Prep Set for Illumina (Set11, New England Biolab®, Germany) on the platform NextSeq 500 platform (Illumina, USA). CDNA relative expression level was assessed by the fold change using ΔΔCt method.
M0s1 / M0s2 = 2-∆∆Ct,
Where M0s1 and M0s2 are initial amounts of cDNA in samples s1 and s2, ΔΔCt = (Cts1 - Ctnorm1) - (Cts2 - Ct norm2), Ct is an amplification cycle value at the point of intersection of the kinetic curve and accumulation of amplificated product with a line of fluorescence threshold level, which is determined automatically by the software StepOnePlus amplificator; Cts1 and Cts2 is the cycle threshold amplification value of cDNA which was synthesized for sncRNA analysis in two compared s1 and s2 samples; Ctnorm1 and Ctnorm2 is the value of the threshold amplification cycle of cDNA for standardising endogenous sncRNA in s1 and s2 samples. The control sample was culture medium without an embryo; to standardize the data of the polymer chain reaction we chose sncRNA hsa- piR023338 | gb | DQ601914 because of its stable expression in all samples (n = 49).
All samples of the embryo culture media included in the study were obtained from 39 patients with tubal factor infertility. ICSI program was performed in 35 patients and IVF program was conducted in 4 patients using the standard protocol of ovarian stimulation, commenced on day 2-3 of the menstrual cycle. We used gonadotropin releasing hormone antagonist and gonadotropins. In patients included in the study we analysed their medical history, clinical parameters, hormonal status, and the results of previous IVF programs (pregnancy+ / pregnancy-). All women were examined in accordance with the order of the Ministry of Health of the Russian Federation No. 107n.
The inclusion criteria were as follows: patients aged 20-37 years with normal ovarian reserve, tubal factor infertility, regular menstrual cycle. We did not include patients with contraindications for IVF or ICSI, in accordance with the order of the Russian Federation Ministry of Health №107n.
All patients were divided into three groups depending on the IVF program result (Table 2).

Group I: 25 patients who had ovarian stimulation and embryo transfer in stimulated cycle with negative blood pregnancy test.
Group II: 14 patients who had ovarian stimulation and embryo transfer in stimulated cycle with positive blood pregnancy test.
Group III: 4 patients from group I who had implantation failure in the previous stimulated cycle, and they had frozen-thawed (FT) embryo transfer with a positive result. All embryos were thawed at the blastocyst stage. The endometrium was prepared with the exogenous administration of oral micronized estradiol forms and progesterone on day 5-6 of the menstrual cycle. The dosage of estradiol was chosen on an individual basis (4-6 mg/day). FT embryo transfer was performed on day 19-20 of the menstrual cycle. Endometrium thickness on the day of embryo transfer was 9-12 mm.
Evaluation of age and anthropometric characteristics in studied groups revealed no statistically significant differences (Table 2). The groups were matched according to these parameters. All patients were selected in such a way as to minimise the influence of interfering factors on the IVF program results. When analysing the menstrual cycle, ovarian reserve, reproductive history and the history of gynaecological diseases in patients enrolled in the current study no statistically significant differences were found.
In stimulated cycles, human chorionic gonadotrophin (HCG) injection was administered to patients from groups I and II to trigger final oocyte maturation. The trigger was administered when the follicles reached ≥17 mm in diameter. The mature oocytes were retrieved after 34-36 hours of HCG injection. The collected matured oocytes were fertilised in-vitro using IVF (4 patients) and ICSI method (35 patients). Progesterone vaginal suppository or dydrogesterone was started after oocyte retrieval on a daily basis and continued until the result of the first blood pregnancy test in patients from groups I-III.
All embryos were cultivated in the individual microdrops in the incubator. Embryo transfer was performed on day 5 after oocyte retrieval under the ultrasound guide.
Data processing
Scripts written in R and Rstudio program were used for data processing [14, 15]. The Shapiro-Wilk test was performed to test if the analyzed parameters were normally distributed. The χ2 test was used for comparing categorical variables. The ANOVA test was applied for the analysis of three groups of normally distributed parameters. Also, Mann-Whitney U-test was used for the pairwise comparison of the non-normally distributed parameters. Absolute numbers (N) and percentages of the total number of patients in group (P) in the N (P%) format were used to describe categorical binary data. The arithmetic mean (M) and standard deviation (SD) in the M (SD) format were used to evaluate the normally distributed quantitative data. A non-normally distributed parameters were described as medians (Me) and quartiles (Q1 and Q3) in the Me (Q1; Q3) format. The Spearman correlation analysis was performed since both quantitative and qualitative data were analyzed. The 95% confidence interval for the correlation coefficients was determined using the Fisher transform strategy. Thresholds for the p-value statistical significance was 0.05. The p-value was specified in the p-value < 0.001 format if it was less than 0.001. In addition, the morphological characteristics of the embryos and the obtained experimental data were analyzed using Partial Least Squares Discriminant Analysis (PLS-DA) [16].
Results and Discussion
Firstly, a deep sequencing was performed to identify microRNA и piwiRNA inside the blastocyst cavity and its culture medium on day 4 after fertilization. It was found that piwiRNAs had a wider spectrum (132 and 128 subtypes in blastocoele fluid and embryo culture medium, respectively) in comparison with microRNAs (49 and 36 subtypes in blastocoele fluid and embryo culture medium, respectively). Among them, 73.5% of microRNAs and 34.7% of piwiRNAs are secreted by the embryo into both the blastocoele fluid and embryo culture medium. All the microRNAs secreted by the embryo into the culture medium were detected in the blastocoele fluid. As for piwiRNAs, 33.7% were uniquely expressed into the blastocoele fluid and 31.6% into the embryo culture medium. To analyze the microRNAs and piwiRNAs using quantitative polymerase chain reaction with reverse transcription (RT-PCR), we selected the molecules with the number of reads not less than 25. In a pilot experiment with 24 samples we standardized RT-PCR using specific primers for the selected microRNAs and piwiRNAs. Significant signals ( Ct <35 cycles) and specific reaction products were obtained for five microRNAs and two piwiRNAs (hsa-let-7b-5p, hsa-let-7i-5p, hsa-let-7c-5p, hsa-miR-92a-3p, hsa-miR-143-3p, hsa_piR_020401|gb|DQ598029|Homo, hsa_piR_023338|gb|DQ601914|Homo).
These sncRNAs were analyzed by quantitative PCR in real time in all RNA samples (n = 49), identified in embryo culture medium. PLS-DA method of RT-PCR data was performed to classify 49 embryo culture media samples according to the embryo quality and the degree of the sncRNA expression profiles similarity.
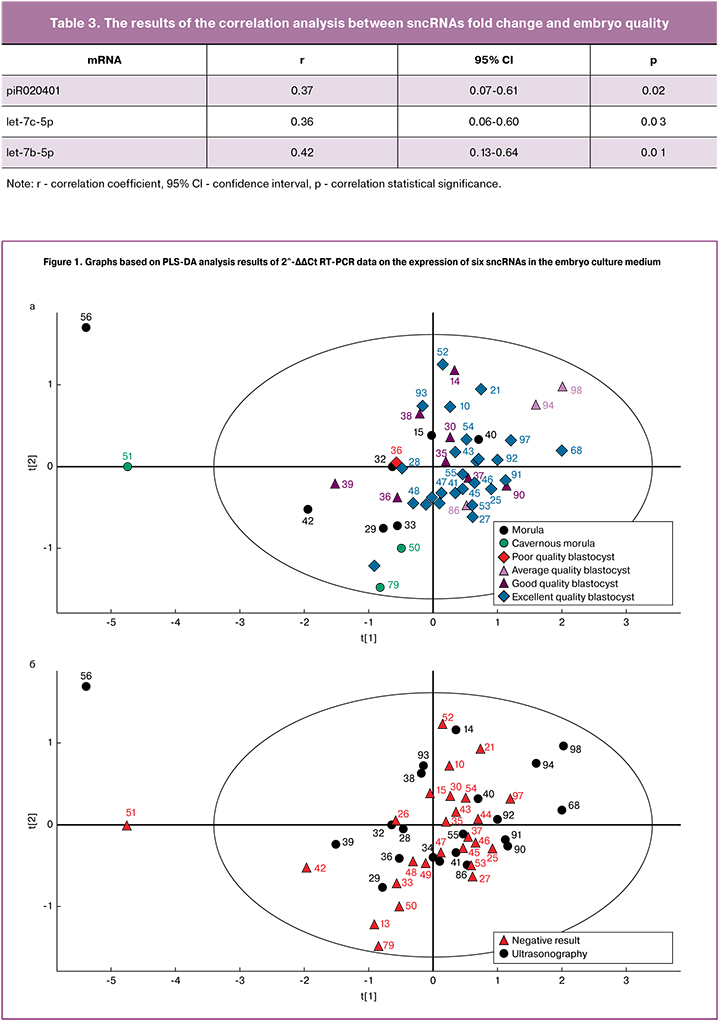
The samples were ranked beginning from embryos of the worst quality and finishing with the best quality embryos; line “ poor quality morula-cavernous morula-blastocyst, average quality blastocyst, good quality blastocyst, excellent quality blastocyst” has been transformed into a series of “0, 1, 2, 3, 4, 5”. PLS-DA model was designed according to the results of RT-PCR data in the following way 2^-ΔΔCt for each sncRNA studied in one sample. Figure 1 shows the graph based on PLS-DA analysis results. The points corresponding to the embryos of excellent, good and average quality are grouped together, preferably in the first and forth quadrant of the graph, and most of the points corresponding to morulas and cavernous morulas, formed a cluster in the third quadrant (Fig. 1a). Thus, embryo development stage depends on sncRNA expression profile in embryo culture media and can be assessed with the help of these biomarkers.
Contribution of six sncRNAs to the possibility of embryos classification depending on their quality is characterised by variable importance in projection (VIP). According to the resulting VIP values, four sncRNAs are crucial in establishing embryo quality: piR020401 (VIP=1.46027), let-7c-5p (VIP=1.17416), let-7b-5p (VIP=0.994657), let-7i-5p (VIP=0.942665), and these molecules have minimum significance - miR-143-5р (VIP=0.623108) and miR-92a-3p (VIP=0.471951).
The contribution of piR020401, let-7c-5p and let-7b-5p to the embryo quality is confirmed by Spearman correlation analysis. The ranking of 49 samples was performed the same way as for PLS-DA model (see above). A statistically significant correlation was found between the expression level of particular sncRNAs in the embryo culture medium and the embryo quality (Table 3).
The relationship between embryo morphological characteristics and embryo quality is shown in box-diagram (Fig. 2), where the differences in sncRNA expression profile in different embryo groups are clearly demonstrated. SncRNA expression profile analysis in the compared groups revealed statistically significant differences using Mann – Whitney method (Table 4).

When we applied the results of embryo implantation (implantation+/-) on the model designed to classify embryos according to their quality, we did not find any noticeable points clustering which may correspond to positive and negative result of implantation (Fig. 1b). This confirms the fact that embryo implantation potential depends on gamete quality and other “parental” factors. According to some studies, patients with tubal factor infertility may have reduced number and less sensitive estrogen and progesterone receptors in the endometrium [17]. As a result, endometrial receptivity is impaired, which may cause IVF failure. It is worth noting that inadequate endometrium receptivity is responsible for almost two thirds of implantation failures [18].
In addition, according to the American Society for Reproductive Medicine (ASRM), a higher incidence of implantation, pregnancy and live birth has been reported in patients who underwent FT embryo transfer [19]. We can suggest that due to this fact 8% of patients in this study became pregnant after FT embryo transfer cycles after previous failure in stimulation cycles (№ embryos, 56, 94, 68) (Fig. 1). The development of cryopreservation technology makes it possible to maintain embryo quality and their implantation potential which is the same as in fresh embryos [20]. Every year more and more data appear in favor of FT embryo transfer performed next menstrual cycle without hormonal stimulation and any negative effects on the endometrium. This strategy can be applied since cycle segmentation allows the prevention of ovary hyperstimulation syndrome. Embryo can also be transferred in a more physiologic uterine environment, without negative influence on endometrium receptivity [21]. It is shown that a premature rise in progesterone during ovulation stimulation was accompanied by premature endometrium secretory transformation and genes expression disorder responsible for implantation (HOXA 10 receptor, Leukemia inhibitory factor receptor and others) [22].
Probably, due to these factors, embryos from A and B quality do not always successfully implant. In addition, the implantation potential is determined by male infertility factor, the presence/absence of assisted hatching and others. On the other hand, the embryos of worse quality in some cases can lead to a successful pregnancy. Thus, we cannot use only morphological criteria to select embryos for transfer, it is necessary to consider different factors [23].
Conclusion
In recent years, attention of scientists has been drawn to the sncRNAs identified in embryo culture media during embryogenesis. The results of the current study show that these molecules make it possible to differentiate the stages of embryo development. To understand the contribution of identified sncRNAs to the development and implantation potential, further research of sncRNA expression profile at morula stage is required, specifically: 1) its differentiation into the blastocyst; 2) degeneration; 3) arrested development on the 5th day after fertilization. Thus, the algorithm to observe embryos at different development stages can be optimized. Study of piR020401, let-7c-5p, let-7b-5p and miR-92a-3p expression profile in embryo culture medium is a non-invasive and informative method for the best embryo selection. This technique meets all criteria required for the most promising and innovative diagnostic tools and allows to standardize IVF programs, taking into account associated factors.
Acknowledgments: the study was conducted for the account of financing the state project “Improving the assisted reproductive technology programs in the application of innovative high-tech (embryological, cellular, immunological, molecular genetic) techniques”. Registration number: AAAA-A18-118053190022-8
References
- Сухих Г.Т., Назаренко Т.А., ред. Бесплодный брак. Современные подходы к диагностике и лечению. М.: ГЭОТАР-Медиа; 2010 518c. .[Sukhikh, G.T., Nazarenko, T.A., ed. Infertile marriage. Modern approaches to diagnosis and treatment. M.: GEOTAR-Media; 2010 518c. (in Russian)]
- Корсак В.С., Смирнова А.А., Шурыгина О.В. ВРТ в России. Отчет за 2013 год. Регистр ВРТ. СПб.: Российская Ассоциация Репродукции Человека; 2015: 26-9. [Korsak V. S., Smirnova A. A., Shurygina O. V. ART in Russia. Report for 2013. ART Register: Russian Association of Human Reproduction; 2015: 26-9.(in Russian)]
- Gardner D.K., Balaban B. Assessment of human embryo development using morpho-logical criteria in an era of time-lapse, algorithms and ‘OMICS’: is looking good still important? Mol. Hum. Reprod. 2016; 22(10): 704-18.
- Rocha J.C., Passalia F., Matos F.D., Maserati M.P. Jr., Alves M.F., Almeida T.G. et al. Methods for assessing the quality of mammalian embryos: How far we are from the gold standard? JBRA Assist. Reprod. 2016; 20(3): 150-8.
- Кулакова Е.В., Калинина Е.А., Трофимов Д.Ю., Макарова Н.П., Хечумян Л.Р., Дударова А.Х. Вспомогательные репродуктивные технологии у супружеских пар с высоким риском генетических нарушений. Преимплантационный генетический скрининг. Акушерство и гинекология. 2017; 8: 21-7. [ Kulakova E. V., Kalinina E. A., Trofimov D. Yu., Makarova N. P. Khechumyan L. R., Dudarova, A. H. Assisted reproductive technologies for married couples with a high risk of genetic disorders. Preimplantation genetic screening. Obstetrics and gynecology. 2017; 8: 21-7. (in Russian)]
- Драпкина Ю.С., Тимофеева А.В., Чаговец В.В., Кононихин А.С., Франкевич В.Е., Калинина Е.А. Применение омиксных технологий в решении проблем репродуктивной медицины. Акушерство и гинекология. 2018; 9: 24-32. [ Drapkina J. S., Timofeev A. V., Chagovets V. V., Kononikhin A. S., She, V. E., Kalinina E. A. The use of omics technologies in reproductive medicine. Obstetrics and gynecology. 2018; 9: 24-32. (in Russian)]
- Kropp J., Salih S., Khatib H. Expression of microRNAs in bovine and human pre-implantation embryo culture media. Front. Genet. 2014; 5: 91.
- Gilchrist G., Tscherner A., Nalpathamkalam T., Merico D., LaMarre J. MicroRNA expression during bovine oocyte maturation and fertilization. Int. J. Mol. Sci. 2016; 17(3): 396.
- Houwing S., Kamminga L., Berezikov E., Cronembold D., Girard A., Elst H. et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007; 129(1): 69-82.
- Rosenbluth E., Shelton D., Sparks A., Devor E., Christenson L., Van Voorhis B. MicroRNA expression in the human blastocyst. Fertil. Steril. 2013; 99(3): 855-61. e3.
- Roth L.W., McCallie B., Alvero R., Schoolcraft W.B., Minjarez D., Katz-Jaffe M. Altered microRNA and gene expression in the follicular fluid of women with polycystic ovary syndrome. J. Assist. Reprod. Genet. 2014; 31(3): 355-62.
- Capalbo A., Ubaldi F.M., Cimadomo D., Noli L., Khalaf Y., Farcomeni A. et al. MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil. Steril. 2016; 105(1): 225-35. e1-3.
- Liang J., Wang S., Wang Z. Role of microRNAs in embryo implantation. Reprod Biol. Endocrinol. 2017; 15(1): 90.
- R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. Available at: https://www.R-project.org/ и программу RStudio
- RStudio Team. RStudio: Integrated development for R. RStudio. Boston, MA: RStudio Inc.; 2016. Available at: http://www.rstudio.com/
- Wold S., Sjöström M., Eriksson L. PLS-regression: a basic tool of chemometrics. Chemometr. Intell. Lab. Syst. 2001; 58(2): 109-30.
- Ruiz-Alonso M., Blesa D., Díaz-Gimeno P., Gómez E., Fernández-Sánchez M., Carranza F. et al. The endometrial receptivity array for diagnosis and personalized embryo transfer as a treatment for patients with repeated implantation failure. Fertil. Steril. 2013; 100(3): 818-24.
- Altmäe S., Koel M., Võsa U., Adler P., Suhorutšenko M., Laisk-Podar T. et al. Meta-signature of human endometrial receptivity: a meta-analysis and validation study of transcriptomic biomarkers. Sci. Rep. 2017; 7(1): 10077.
- Practice Committee of the American Society for Reproductive Medicine. Performing the embryo transfer: a guideline. 2013.
- Evans J., Hannan N.J., Edgell T.A., Vollenhoven B.J., Lutjen P.J., Osianlis T. et al. Fresh versus frozen embryo transfer: backing clinical decisions with scientific and clinical evidence. Hum. Reprod. Update. 2014; 20(6): 808-21.
- Roque M., Valle M., Guimarães F., Sampaio M., Geber S. Freeze-all policy: fresh vs. frozen-thawed embryo transfer. Fertil. Steril. 2015; 103(5): 1190-3.
- Bashiri A., Halper K.I., Orvieto R. Recurrent implantation failure-update overview on etiology, diagnosis, treatment and future directions. Reprod. Biol. Endocrinol. 2018; 16(1): 121.
- Смольникова В.Ю., Калинина Е.А., Краснощока О.Е, Донников А.Е., Бурменская О.В., Трофимов Д.Ю., Сухих Г.Т. Возможности неинвазивной оценки состояния ооцита и эмбриона при проведении программ ВРТ по профилю экспрессии мРНК факторов роста в фолликулярной жидкости. Акушерство и гинекология. 2014; 9: 36-43. [Smolnikova V.Yu., Kalinina E.A., Krasnoshchoka O.E., Donnikov A.E., Burmenskaya O.E., Trofimov D.Yu., Sukhikh G.T. Possibilities for noninvasive oocyte and embryo evaluation when implementing assisted reproductive technology programs for follicular-fluid growth factor mRNA expression. Obstetrics and gynecology. 2014; 9: 36-43. (in Russian)]
Received 20.01.2019
Accepted 22.02.2019
About the Authors
Timofeeva Angelika Vladimirovna, PhD, Chief of the Laboratory of Transcriptomic at the Department of Systems Biology in ReproductionKulakov National Medical Research Center of Obstetrics, Gynecology, and Perinatology, Ministry of Health of Russia.
117997, Russia, Moscow, Ac. Oparina 4. Tel. +7(495) 438-13-41. E-mail avtimofeeva28@gmail.com
Kalinina Elena Anatolievna, MD, Associate Professor, Chief of the Department of Reproductive Health named after professor Leonov
Kulakov National Medical Research Center of Obstetrics, Gynecology, and Perinatology, Ministry of Health of Russia.
117997, Russia, Moscow, Ac. Oparina 4. Tel. +7(495) 438-13-41 E-mail: e_kalinina@oparina4.ru
Drapkina Yulia Sergeevna, PhD student at the Department of Reproductive Health named after professor Leonov
Kulakov National Medical Research Center of Obstetrics, Gynecology, and Perinatology, Ministry of Health of Russia.
117997, Russia, Moscow, Ac. Oparina 4. Tel. +79169500745 E-mail: julia.drapkina@gmail.com
Chagovets Vitaliy Viktorovich, PhD, Senior Researcher of the Laboratory of Proteomics and Metabolomics in Human Reproduction at the Department of Systems Biology
in Reproduction Kulakov National Medical Research Center of Obstetrics, Gynecology, and Perinatology, Ministry of Health of Russia.
117997, Russia, Moscow, Ac. Oparina 4. Tel. 8 (926) 562-65-90. E-mail: vvchagovets@gmail.com
Makarova Nataliya Petrovna, PhD, senior embriologist at the Department of Reproductive Health named after professor Leonov
Kulakov National Medical Research Center of Obstetrics, Gynecology, and Perinatology, Ministry of Health of Russia.
117997, Russia, Moscow, Ac. Oparina 4. Tel. + 7(495) 438-13-41. E-mail: npmakarova@gmail.com
Sukhikh Gennady Tikhоnovich, MD, PhD, Professor, Academician of Russian Academy of Sciences, Director of Kulakov National Medical Research Center of Obstetrics, Gynecology, and Perinatology, Ministry of Health of Russia 17997, Russia, Moscow, Ac. Oparina 4. Tel. +7(495) 438-18-00. E-mail: gtsukhih@mail.ru
For citations: A.V. Timofeeva, E.A. Kalinina, Yu.S. Drapkina, V.V. Chagovets, N.P. Makarova, G.T. Sukhikh Embryo quality assessment by the small noncoding RNA expression profile in an embryo culture medium in assisted reproductive technology programs. Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2019; (6):79-86 (in Russian).
http://dx.doi.org/10.18565/aig.2019.6.79-86



