Molecular genetic determinants of adenomyosis
Altukhova O.B., Radzinsky V.E., Sirotina S.S., Efremova O.A., Batlutskaya I.V., Orlova V.S., Churnosov M.I.
Relevance: Adenomyosis is a common disease of the female reproductive system that affects 15–45% of women. One important aspect of the pathogenesis of adenomyosis is the disruption of apoptosis and immune processes, which are caused by an imbalance of growth factors (VEGF, EGF, TGFβ1, IGF1, and FGFR2). Polymorphisms in growth factor genes may influence their expression and therefore play a role in the pathophysiology of adenomyosis.
Objective: To evaluate the involvement of polymorphic loci in growth factor genes in the development of adenomyosis.
Materials and methods: This study included 102 patients with adenomyosis and 778 control women. Five polymorphic growth factor gene loci were selected for the study: rs4444903 EGF c.-382A>G and rs6214 IGF1 c.*2716G>A, rs2981582 FGFR2 c.109+906T>C, rs833061 VEGF c.-958C>T, rs1800469 TGFb1 c .-1347 T>S. The analysis was performed by real-time PCR. The APSampler program was used to assess inter-locus interactions (https://sourceforge.net/projects/apsampler/).
Results: The study results showed that the genotype GG IGF1 rs6214 (OR=2.64, p=0.01) should be considered a risk factor for the development of adenomyosis, as well as the combination of polymorphic variants G rs6214 IGF1, C rs1800469 TGFb1, A rs4444903 EGF, and T rs833061 VEGF (OR=1.88, pperm=0.0021), and the alleles G rs6214 IGF1, C rs1800469 TGFb1, T rs833061 VEGF, and C rs2981582 FGFR2 (OR=1.71, pperm=0.003).
Conclusion: The gene combinations rs4444903 EGF, rs6214 IGF1, rs2981582 FGFR2, rs833061 VEGF, and rs1800469 TGFb1 are associated with the development of adenomyosis. These results indicate the importance of interlocus interactions between growth factor genes in the development of adenomyosis. In the future, the obtained data could be used in practical medicine.
Authors' contributions: Churnosov M.I., Altukhova O.B., Radzinsky V.E. – conception and design of the study; Altukhova O.B., Churnosov M.I., Batlutskaya I.V. – material collection and processing; Sirotina S.S., Efremova O.A. – drafting of the manuscript; Churnosov M.I., Altukhova O.B., Orlova V.S. – editing of the manuscript.
Conflicts of interest: The authors have no conflicts of interest to declare.
Funding: There was no funding for this study.
Ethical Approval: The study was reviewed and approved by the Research Ethics Committee of the Belgorod State National Research University (Ref. No: 4 of October 21, 2019).
Patient Consent for Publication: All patients provided informed consent for the publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available upon request from the corresponding author after approval from the principal investigator.
For citation: Altukhova O.B., Radzinsky V.E., Sirotina S.S., Efremova O.A., Batlutskaya I.V.,
Orlova V.S., Churnosov M.I. Molecular genetic determinants of adenomyosis.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2024; (4): 69-74 (in Russian)
https://dx.doi.org/10.18565/aig.2023.272
Keywords
Adenomyosis is a common gynecological disease characterized by benign proliferation and chronic inflammation. It has significant social implications because it reduces the quality of life of women. The disease is known for its pronounced clinical manifestations and tendency to relapse even after treatment [1, 2]. The symptoms of adenomyosis include frequent aching pain in the pelvic area, irritable bowel syndrome, dysmenorrhea, painful ovulation, and dyspareunia. Despite its high prevalence among women of reproductive age, the pathogenesis of adenomyosis is not yet fully understood [3, 4].
The development of adenomyosis is attributed to hormonal imbalances, dysfunction in stem cell activity and growth factors, as well as genetic and epigenetic abnormalities [5–7]. Genetic determinants, such as genes encoding growth factors, play a role in adenomyosis formation. These genes are directly or indirectly involved in processes such as proliferation, differentiation, and cell migration and exhibit immunosuppressive properties [8–10].
This study aimed to evaluate the involvement of polymorphic loci in growth factor genes in the development of adenomyosis.
Materials and methods
To conduct a molecular genetic study, patients with adenomyosis (study group) and women from the control group were selected from the regional clinical hospital in the city of Belgorod. The study included 880 women of Russian nationality living in the Belgorod region, who were not related to each other and agreed to participate in the study. Patients with adenomyosis were included in the study group after the diagnosis of the disease was established and confirmed using clinical, clinical, instrumental, and clinical laboratory investigations. Exclusion criteria from the study group were malignant diseases of the female reproductive system and pregnancy. Women in the control group were selected from among women without reproductive system diseases. Thus, according to the above criteria, the sample for this study included 102 patients in the study group and 778 women in the control group. The study was approved by the Regional Ethics Committee of Belgorod State National Research University (Protocol 4 of October 21, 2019).
When planning the study, the following DNA markers were selected: rs4444903 EGF c.-382A>G (epidermal growth factor), rs6214 IGF1 c.*2716G>A (insulin-like growth factor 1), rs2981582 FGFR2 c.109+906T>C (factor receptor fibroblast growth), rs833061 VEGF c.-958C>T (vascular endothelial growth factor), rs1800469 TGFb1 c.-1347 T>C (transforming growth factor beta 1) [11]. For the polymerase chain reaction (PCR), DNA was isolated from peripheral blood leukocytes using phenol-chloroform extraction. Molecular genetic loci were analyzed using real-time PCR and oligonucleotide primers and probes (Syntol LLC, Russia). During molecular genetic analysis, some DNA samples were removed from the study because the level of relative fluorescence during genotyping did not allow the genotype to be determined with sufficient accuracy.
Statistical methods
The normality of the distribution was tested using the Shapiro-Wilk test. Continuous variables showing normal distribution (age, body mass index (BMI)) were expressed as mean (M) and standard deviation (SD) and compared using Student’s t-test. Allele and genotype frequencies between the study and control groups were compared using the χ2 test with Yates correction for continuity. Calculations were performed using 2×2 contingency tables [12].
Associations of combinations of alleles of the analyzed genes with the development of adenomyosis were assessed using the APSampler program (https://sourceforge.net/projects/apsampler/) using the Monte Carlo Markov chain method and Bayesian nonparametric statistics, calculating the odds ratio (OR) and its 95% confidence interval (95% CI). To determine the effects of false-positive results in multiple comparisons, a permutation test (pperm) was used. Pperm <0.05 was taken as a statistically significant level [13]. Statistical analysis was performed using STATISTICA for Windows 10.0 and Microsoft Excel 2016 software.
Results and discussion
Clinical and demographic characteristics of the study groups are presented in Table 1.
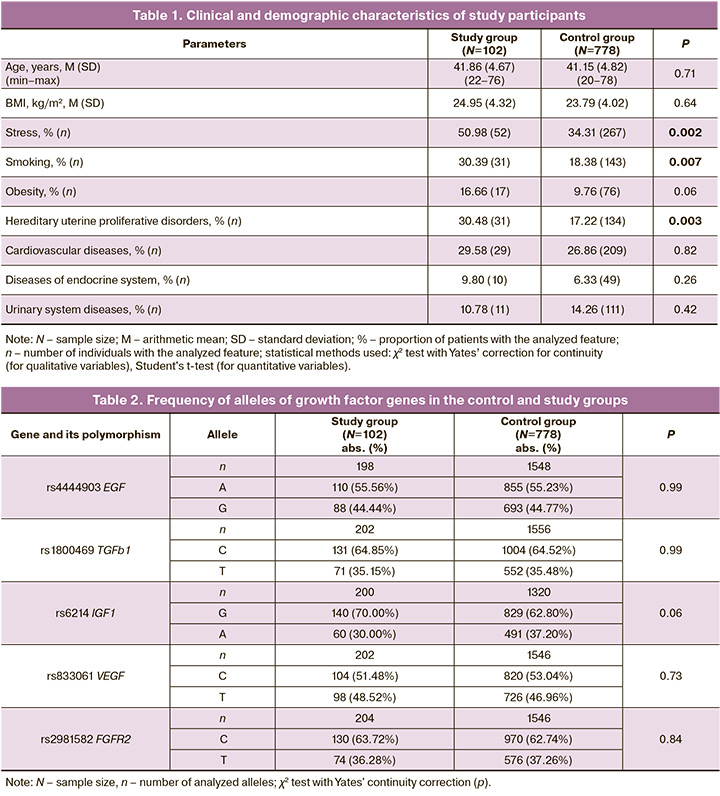
The mean age of the patients was 41.86 (4.67) years in the study group and 41.15 (4.82) years in the control group (p=0.71). Among the comorbidities, cardiovascular (29.58% and 26.86%), endocrine (9.80% and 6.33%), and genitourinary (10.78% and 14.26%) diseases were more common in both the study and control groups. There were no statistically significant differences in these parameters between the study groups (p=0.82, p=0.26, and p=0.42, respectively). It should be noted that parameters such as stress, smoking and inheritance of proliferative uterine diseases were significantly different between the study and control groups (p=0.002, p=0.007 and p=0.003, respectively).
Tables 2 and 3 show the results of the statistical analysis of the data obtained by genotyping the polymorphisms of growth factors in the control and study groups.
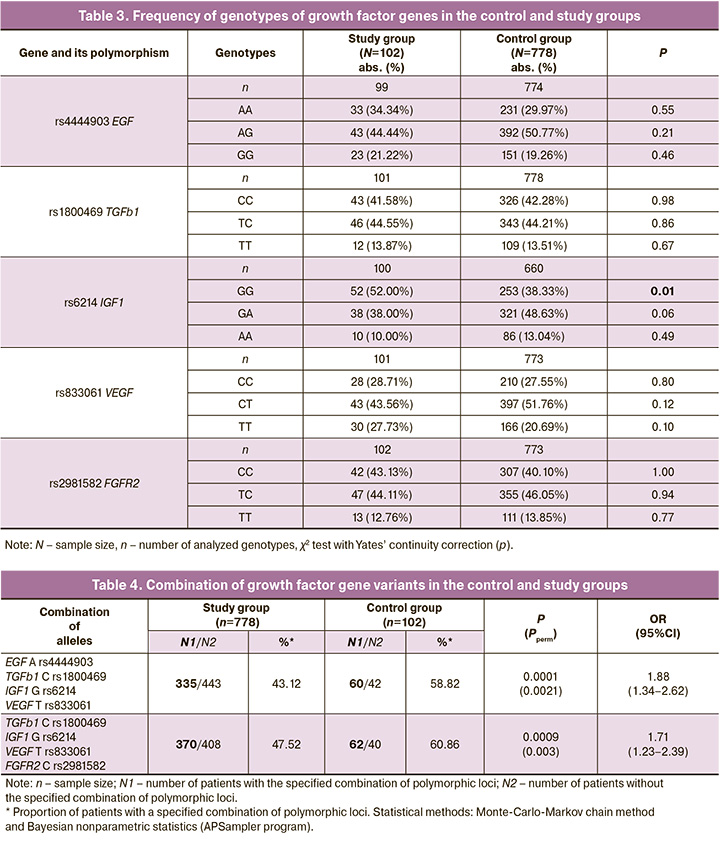
Thus, the GG IGF1 rs6214 genotype in the study group occurred in 52.00% (52/102) of cases and in 38.33 % (253/778) of cases in the control group (χ2=1.74, p=0.01, OR=2.64, 95% CI 1.14–2.65).
According to the literature, IGF1 in physiological concentrations regulates apoptotic processes and the action of steroid hormones, and act as mediators of cell growth, transformation, and differentiation [14].
The polymorphic A locus rs6214 IGF1 causes a decrease in the expression of the corresponding protein, whereas the G allele rs6214 IGF1, which in our study is associated with the risk of developing adenomyosis, has the opposite effect, increasing endometrial proliferation and apoptosis, which can lead to the development of adenomyosis [15].
Risk combinations of growth factor gene alleles for adenomyosis were identified (Table 4). In the study group, the combination of G rs6214 IGF1, C rs1800469 TGFb1, A rs4444903 EGF, and T rs833061 VEGF occurred in 58.82% (60/102) of patients with adenomyosis, and in the control group, in 43.12% (335/778) of women (OR=1.88, pperm=0.0021). In addition, the combination of G rs6214 IGF1, C rs1800469 TGFb1, T rs833061 VEGF, and C rs2981582 FGFR2 was detected in 60.86% (62/102) of patients in the study group and 47.52 % (370/778) of the control group (OR=1.71, pperm=0.003). Established combinations of polymorphic loci of growth factor genes are markers for the risk of developing adenomyosis.
According to literature, the single-nucleotide substitutions we analyzed are localized in the region of modified histones that mark promoters and enhancers in various tissues, A>G rs4444903 EGF, T>C rs2981582 FGFR2, C>T rs833061 VEGF, T>C rs1800469 TGFb1 are located in the region of open chromatin, A>G rs4444903 EGF, rs833061 VEGF, T>C rs1800469 TGFb1 are located in sites that interact with transcription factors, G>A rs6214 IGF1, T>C rs2981582 FGFR2, C>T rs833061 VEGF, T>C rs1800469 TGFb1 are located in regions of DNA regulatory motifs [16]. Thus, the polymorphic genes of growth factors included in this study are important in the body and may be involved in the etiopathogenesis of adenomyosis [17].
The data obtained on the role of polymorphic loci of growth factors in the development of adenomyosis are explained by the medical and biological effects of the protein products of these genes. According to the literature, changes in the production of growth factors stimulate differentiation and ectopic growth of the endometrium, remodeling of the extracellular matrix, and cell migration [18]. Growth factors are also involved in the pathological and physiological neovascularization of the endometrium. Increased production of growth factors in adenomyosis may contribute to its pathophysiology in a manner similar to its carcinogenic effects, causing changes in cellular metabolism, increasing cell invasion, and initiating neoangiogenesis [19, 20].
Conclusion
The gene combinations rs4444903 EGF, rs6214 IGF1, rs2981582 FGFR2, rs833061 VEGF, and rs1800469 TGFb1 are associated with the development of adenomyosis. These results indicate the importance of interlocus interactions between growth factor genes in the development of adenomyosis. In the future, the obtained data could be used in practical medicine.
References
- Chapron C., Marcellin L., Borghese B., Santulli P. Rethinking mechanisms, diagnosis and management of endometriosis. Nat. Rev. Endocrinol. 2019; 15(11): 666-82. https://dx.doi.org/10.1038/s41574-019-0245-z.
- Самойлова А.В., Гунин А.Г., Сидоров А.Е., Денисова Т.Г., Чернышов В.В., Смирнова Т.Л. Современные направления изучения этиологии и патогенеза эндометриоза (обзор литературы). Проблемы репродукции. 2020; 26(5): 118-32. [Samoilova A.V., Gunin A.G., Sidorov A.E., Denisova T.G., Chernyshov V.V., Smirnova T.L. Actual research trends in etiology and pathogenesis of endometriosis (a review). Russian Journal of Human Reproduction. 2020; 26(5): 118-32. (in Russian)]. https://dx.doi.org/10.17116/repro202026051118.
- Taylor H.S., Kotlyar A.M., Flores V.A. Endometriosis is a chronic systemic disease: clinical challenges and novel innovations. Lancet. 2021; 397(10276): 839-52. https://dx.doi.org/10.1016/S0140-6736(21)00389-5.
- Андреев А.Е., Клейменова Т.С., Дробинцева А.О., Полякова В.О., Кветной И.М. Сигнальные молекулы, вовлеченные в образование новых нервных окончаний при эндометриозе (обзор). Научные результаты биомедицинских исследований. 2019; 5(1): 94-107. [Andreev A.E., Kleimenova T.S., Drobintseva A.O., Polyakova V.O., Kvetnoy I.M. Signal molecules involved in the formation of new nerve endings in endometriosis (review). Research Results in Biomedicine. 2019; 5(1): 94-107. (in Russian)]. https://dx.doi.org/10.18413/2313-8955-2019-5-1-0-7.
- Головченко И.О. Генетические детерминанты уровня половых гормонов у больных эндометриозом. Научные результаты биомедицинских исследований. 2023; 9(1): 5-21. [Golovchenko I.O. Genetic determinants of sex hormone levels in endometriosis patients. Research Results in Biomedicine. 2023; 9(1): 5-21. (in Russian)]. https://dx.doi.org/10.18413/2658-6533-2023-9-1-0-1.
- Пономаренко И.В., Полоников А.В., Чурносов М.И. Ассоциация полиморфизма rs4986938 гена ESR2 с развитием гиперплазии эндометрия. Акушерство и гинекология. 2019; 4: 66-72. [Ponomarenko I.V., Polonikov A.V., Churnosov M.I. Association of ESR2 rs4986938 polymorphism with the development of endometrial hyperplasia. Obstetrics and Gynecology. 2019; (4): 66-72. (in Russian)]. https://dx.doi.org/10.18565/aig.2019.4.66-72.
- Алтухова О.Б., Радзинский В.Е., Сиротина С.С., Чурносов М.И. Анализ ассоциации полиморфных вариантов генов рецепторов эстрогенов и прогестерона с развитием генитального эндометриоза. Акушерство и гинекология. 2021; 9: 93-9. [Altukhova O.B., Radzinsky B.E., Sirotina S.S., Churnosov M.I. Analysis of the association between the polymorphic variants of estrogen and progesterone receptor genes and genital endometriosis. Obstetrics and Gynecology. 2021; (9): 93-9. (in Russian)]. https://dx.doi.org/10.18565/aig.2021.9.93-99.
- Алтухова О.Б., Радзинский В.Е., Полякова И.С., Сиротина С.С., Батлуцкая И.В., Орлова В.С., Ефремова О.А., Чурносов М.И. Роль генов хемокинов в развитии внутреннего генитального эндометриоза в сочетании с гиперпластическими процессами эндометрия. Акушерство и гинекология. 2022; 8: 76-84. [Altukhova O.B., Radzinsky V.E., Polyakova I.S., Sirotina S.S., Batlutskaya I.V., Orlova V.S., Efremova O.A., Churnosov M.I. The role of chemokine genes in the development of adenomyosis with concurrent endometrial hyperplastic processes. Obstetrics and Gynecology. 2022; (8): 76-84 (in Russian)]. https://dx.doi.org/10.18565/aig.2022.8.76-84.
- Пономаренко И.В., Полоников А.В., Чурносов М.И. Молекулярные механизмы и факторы риска развития эндометриоза. Акушерство и гинекология. 2019; 3: 26-31. [Ponomarenko I.V., Polonikov A.V., Churnosov M.I. Molecular mechanisms of and risk factors for endometriosis. Obstetrics and Gynecology. 2019; (3): 26-31. (in Russian)]. https://dx.doi.org/10.18565/aig.2019.3.26-31.
- Méar L., Herr M., Fauconnier A., Pineau C., Vialard F. Polymorphisms and endometriosis: a systematic review and meta-analyses. Hum. Reprod. Update. 2020; 26(1): 73-102. https://dx.doi.org/10.1093/humupd/dmz034.
- Kim H., Ku S.Y., Kim S.H., Choi Y.M., Kim J.G. Association between endometriosis and polymorphisms in insulin-like growth factor binding protein genes in Korean women. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012; 162(1): 96-101. https://dx.doi.org/10.1016/j.ejogrb.2012.01.022.
- Пономаренко И.В. Использование метода Multifactor Dimensionality Reduction (MDR) и его модификаций для анализа ген-генных и генно-средовых взаимодействий при генетико-эпидемиологических исследованиях (обзор). Научные результаты биомедицинских исследований. 2019; 5(1): 4-21. [Ponomarenko I.V. Using the method of Multifactor Dimensionality Reduction (MDR) and its modifications for analysis of gene-gene and gene-environment interactions in genetic-epidemiological studies (review). Research Results in Biomedicine. 2019; 5(1): 4-21. (in Russian)]. https://dx.doi.org/10.18413/2313-8955-2019-5-1-0-1.
- Радзинский В.Е., Алтухова О.Б. Молекулярно-генетические детерминанты бесплодия при генитальном эндометриозе. Научные результаты биомедицинских исследований. 2018; 4(3): 28-37. [Radzinsky V.E., Altuchova O.B. Molecular-genetic determinants of infertility in genital endometryosis. Research Results in Biomedicine. 2018; 4(3): 28-37. (in Russian)]. https://dx.doi.org/10.18413/2313-8955-2018-4-3-0-3.
- Hossein Razi M., Eftekhar M., Ghasemi N., Hasan Sheikhha M., Dehghani Firoozabadi A. Expression levels of circulatory mir-185-5p, vascular endothelial growth factor, and platelet-derived growth factor target genes in endometriosis. Int. J. Reprod. Biomed. 2020; 18(5): 347-58. https://dx.doi.org/10.18502/ijrm.v13i5.7155.
- Xu G.P., Chen W.X., Xie W.Y., Wu L.F. The association between IGF1 Gene 3'-UTR polymorphisms and cancer risk: A Meta-analysis. Medicine (Baltimore). 2018; 97(51): e13829. https://dx.doi.org/10.1097/MD.0000000000013829.
- Rahmioglu N., Mortlock S., Ghiasi M., Møller P.L., Stefansdottir L., Galarneau G. et al. The genetic basis of endometriosis and comorbidity with other pain and inflammatory conditions. Nat. Genet. 2023; 55(3): 423-36. https://dx.doi.org/10.1038/s41588-023-01323-z.
- Yilmaz B.D., Bulun S.E. Endometriosis and nuclear receptors. Hum. Reprod. Update. 2019; 25(4): 473-85. https://dx.doi.org/10.1093/humupd/dmz005.
- Smolarz B., Szyłło K., Romanowicz H. Endometriosis: epidemiology, classification, pathogenesis, treatment and genetics (review of literature). Int. J. Mol. Sci. 2021; 22(19): 10554. https://dx.doi.org/10.3390/ijms221910554.
- Vlasova-St. Louis I., Bohjanen P.R. Post-transcriptional regulation of cytokine and growth factor signaling in cancer. Cytokine Growth Factor Rev. 2017; 33: 83-93. https://dx.doi.org/10.1016/j.cytogfr.2016.11.004.
- Gross S.M., Rotwein P. Quantification of growth factor signaling and pathway cross talk by live-cell imaging. Am. J. Physiol. Cell Physiol. 2017; 312(3): C328-C340. https://dx.doi.org/10.1152/ajpcell.00312.2016.
Received 20.11.2023
Accepted 28.03.2024
About the Authors
Oksana B. Altukhova, Dr. Med. Sci., Professor of the Department of Obstetrics and Gynecology, Medical Institute, Belgorod State National Research University, 308015, Russia, Belgorod, Victory str., 85, +7(4722)30-13-83, kristalinka@yandex.ruViktor E. Radzinsky, Dr. Med. Sci., Professor, Merited Scholar of the Russian Federation, Academician of the International Academy of Sciences of the Higher School,
Head of the Department of Obstetrics and Gynecology, Faculty of Medicine, Peoples’ Friendship University of Russia, 117198, Russia, Moscow, Miklukho-Maklaya str., 6,
+7(495)360-46-69, radzinskiy-ve@rudn.ru
Svetlana S. Sirotina, PhD (Bio), Associate Professor at the Department of Biomedical Disciplines, Belgorod State National Research University, 308015, Russia, Belgorod, Victory str., 85, +7(4722)30-13-83, sirotina@bsu.edu.ru, https://orcid.org/0000-0002-4163-7863
Mikhail I. Churnosov, Dr. Med. Sci., Professor, Head of the Department of Biomedical Disciplines of the Medical Institute, Belgorod State National Research University, 308015, Russia, Belgorod, Victory str., 85, +7(4722)30-13-83, churnosov@bsu.edu.ru, https://orcid.org/0000-0003-1254-6134
Valentina S. Orlova, Dr. Med. Sci., Professor at the Department of Obstetrics and Gynecology of the Medical Institute, Belgorod State National Research University,
308015, Russia, Belgorod, Victory str., 85, +7(4722)30-13-83, orlova@bsu.edu.ru, https://orcid.org /0000-0003-3882-9191
Irina V. Batlutskaya, Dr. Bio. Sci., Associate Professor, Head of the Department of Biotechnology and Microbiology, Belgorod State National Research University,
308015, Russia, Belgorod, Victory str., 85, +7(4722)30-13-83, bat@bsu.edu.ru, https://orcid.org/0000-0003-0068-6586
Olga A. Efremova, Dr. Med. Sci., Associate Professor, Head of the Department of Faculty Therapy of the Medical Institute, Belgorod State National Research University, 308015, Russia, Belgorod, Victory str., 85, +7(4722)30-13-83, efremova@bsu.edu.ru, https://orcid.org/0000-0003-4967-2556
Corresponding author: Svetlana S. Sirotina, sirotina@bsu.edu.ru



