Genotype complexes of some cytokines in the constitutional predisposition of Russian women of Caucasian origin to the development of uterine leiomyoma
Konenkov V.I., Koroleva E.G., Prokofiev V.F., Shevchenko A.V., Timofeeva Yu.S., Aidagulova S.V., Marinkin I.O.
Objective: This study aimed to compare the distribution of single nucleotide polymorphisms (SNP) in the regulatory regions of proinflammatory and anti-inflammatory cytokine genes between patients with uterine leiomyoma (UL) and healthy women in the Caucasian population of Western Siberia. Additionally, this study assessed the prognostic significance of genetic differences.
Materials and methods: An observational genetic case-control study involving 180 women with UL (aged 23-61 years) and 98 healthy women of the same age was conducted using real-time PCR. Ten SNPs of pro-inflammatory cytokine genes were studied: tumor necrosis factor (TNF) TNF-863C/A, TNF-308G/A, TNF-238G/A, interleukins (IL) IL1B-31T/C, IL6-174C/G, IL8-251T/A, IL17-197A/G, and anti-inflammatory cytokines IL4-590C/T, IL10-592A/C, and IL-10-1082A/G. Standard methods of genetic and bioinformatics analysis were used.
Results: A comparative analysis of the distribution frequency of the 10 SNPs of cytokine genes identified 587 statistically significant complex genotypes associated positively or negatively with the development of UL after Bonferroni correction. The strongest positive associations were observed for combinations of genotypes
TNF-308G/G and IL17-197A/A (OR=7.03, p=0.0162), IL10-1082A/G and IL17-197A/A (OR=10.78, p=0.0423), TNF-308G/G, IL6-174G/C, IL10-592C/C and IL10-1082A/G (OR=12.81, p=0.0414), with prognostic values for UL development ranging from 82 to 100%. Conversely, complex genotypes indicating resistance to UL development (p<0.01) were identified in comparison to the control group, with a prognostic value for a specific woman ranging from 87 to 100%.
Conclusion: The data obtained from this study can be used to establish a set of personalized prognostic indicators for a woman's genetic predisposition or resistance to UL development, even before the disease manifests.
Authors' contributions: Konenkov V.I., Marinkin I.O. – conception and design of the study; Koroleva E.G., Timofeeva Yu.S. – collection and processing of primary material; Timofeeva Yu.S., Aidagulova S.V. – literature data analysis; Shevchenko A.V. – genotyping and immunogenetic analysis; Prokofiev V.F. – statistical analysis; Konenkov V.I., Aidagulova S.V., Prokofiev V.F., Marinkin I.O. – drafting and editing of the manuscript.
Conflicts of interest: The authors have no conflicts of interest regarding the publication of this article.
Funding: The study was conducted within the framework of the State assignment Research Institute of Clinical and Experimental Lymphology – Branch of the Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences of the Ministry of Education and Science of Russia for conducting fundamental scientific research – FWNR-2022-0009 (state registration number of research work 1021060908897-3). Publication of the research results in the open press is permitted.
Ethical Approval: The study was reviewed and approved by the Research Ethics Committee of the RICEL - Branch of IC&G SB RAS (Ref. No: 115 of 24.12.2015) and the Research Ethics Committee of the Novosibirsk SMU of Minzdrav of Russia (Ref. No: 107 of 31.05.2018).
Patient Consent for Publication: All patients provided informed consent for the publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available upon request from the corresponding author after approval from the principal investigator.
For citation: Konenkov V.I., Koroleva E.G., Prokofiev V.F., Shevchenko A.V., Timofeeva Yu.S., Aidagulova S.V., Marinkin I.O. Genotype complexes of some cytokines in the constitutional predisposition of Russian women of Caucasian origin to the development of uterine leiomyoma.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2024; (9): 108-115 (in Russian)
https://dx.doi.org/10.18565/aig.2024.154
Keywords
The etiology of uterine leiomyomas (UL) remains an unsolved problem in modern medicine. This hinders the development of effective drug treatment algorithms for this common disease and often necessitates surgical intervention. The frequency of UL varies from 50% in women of reproductive age to 70–80% among women over 50 years of age. There are numerous concepts regarding the etiopathogenesis of UL, including various factors that contribute to the initiation and progression of multiple monoclonal proliferations of transformed smooth muscle cells in the cervix or body of the uterus. These proliferations can be submucosal, intramural, or subserous. Among the factors involved in the etiopathogenesis of UL, disturbances in the regulation of myocyte mitotic activity, changes in estrogen and progesterone interactions with their receptors, deviations in the concentration of cytokines, chemokines, growth factors, and other signaling molecules that regulate angiogenesis, apoptosis, and extracellular matrix [1–3] remodeling are undoubtedly important.
The search for predictors and triggers of the myomatous process in women of the same or different ethnic groups living in similar social and living conditions is ongoing. Most authors believe that genetic predisposition is a leading factor in the development of UL. One recent review from Johns Hopkins University (2024) lists the most significant genetic abnormalities associated with UL, including mutations in MED12, hyperexpression of HMGA2, fumarate hydratase deficiency (HLRCC), and cytogenetic abnormalities. Epigenetic modifications such as histone acetylation and DNA methylation also play an important role in UL development. Stem cells from patients with UL have a unique DNA methylation pattern compared that with of more differentiated cells. Genome-wide association studies (GWAS) and epidemiological analyses have identified 23 genetic loci associated with earlier age at menarche and UL development. GWAS studies have also explored genetic loci as potential factors contributing to racial differences in UL incidence [4]. However, the results of genome-wide studies, as well as studies on metatranscriptomics and metamethylomics of UL, are not currently being utilized for the development of new diagnostic methods and therapeutic agents [5, 6].
Cytokines, as regulators of cellular activity, also make a significant contribution to the pathogenesis of UL [7, 8]. The implementation of t. Their regulatory properties depend on both the variability of the coding sequences of their genes and quantitative factors determined by non-coding regulatory gene fragments and epigenomic factors [9, 10].
This study aimed to compare the distribution of single nucleotide polymorphisms (SNPs) in the regulatory regions of proinflammatory and anti-inflammatory cytokine genes in patients with uterine leiomyoma (UL) and healthy women in the Caucasian population of Western Siberia, Russia. This study also aimed to assess the prognostic significance of genetic differences.
Materials and methods
This genetic and clinical case-control study included 278 Russian Caucasian women. The study group included 180 women aged 23–61 years (39.7 (6.5)), with fibroids visualized using instrumental examination (D25 according to ICD-10) and verified during subsequent surgical intervention and histological examination. All 180 patients underwent planned myomectomy of single (68%) or multiple (32%) fibroid nodules due to polymenorrhagia and anemia (in half of the observations), as well as for other indications. According to the FIGO classification [11], 82% of the patients had predominant fibroid nodules of types 4 and 5 (intramural), while the remaining 18% had submucosal fibroid nodules (FIGO types 1–2). The most common fibroid nodules were 40–80 mm in diameter, accounting for up to 51% of all observed fibroid nodule size variants. The comorbidities included chronic endometritis, endometrial polyps, intrauterine adhesions, endocervical hyperplasia, and endometrial hyperplasia without atypia.
The control group included 98 healthy women aged 22–61 years (34.0 (6.3)), without UL and other gynecological diseases, who sought a comprehensive examination as part of annual examinations.
The exclusion criteria for both groups were pregnancy, diabetes mellitus, acute inflammatory, acute infectious, and autoimmune diseases, as well as refusal to sign informed consent to participate in the study.
This study was reviewed and approved by the Research Ethics Committee of the RICEL – Branch of the IC&G SB RAS (Ref. No: 115 of 24.12.2015), and the Research Ethics Committee of Novosibirsk SMU, Ministry of Health of Russia (Ref. No: 107 of 31.05.2018).
Genotyping
Human genomic DNA isolated from whole blood leukocytes was analyzed using the "DNA-express-blood" test system (Litex, Russia). Genotyping of single nucleotide polymorphisms (SNP) in tumor necrosis factor (TNF) -863C/A, TNF-308G/A, TNF-238G/A, interleukin (IL) 1B-31T/C, IL4-590 C/T, IL6-174C/G, IL8-251T/A, IL10-592 A/C, IL-10-1082 A/G, and IL17-197A/G was performed by real-time polymerase chain reaction (PCR) using the intercalating dye SYBER GREEN (Litex, Russia) on a DT-96 amplifier. Two amplification reactions using two pairs of allele-specific primers were performed in parallel with the isolated DNA sample. Ready-made tests from the manufacturer were used, and amplification was performed according to the manufacturer’s instructions. The test systems were adapted from the manufacturer for the DT-96 amplifier (DNA Technology, Russia). The results were considered positive if the FAM Ct value was less than 27 and allowed three types of conclusions to be made: homozygotes for allele 1, homozygotes for allele 2, and heterozygotes. In the case of heterozygotes, the difference between the Ct values of the two parallel samples did not exceed 1.5 cycles. Analysis of the specificity of the amplicons and the presence of cross-reactions between the genotypes was not carried out, since commercial test systems were used, and this should have been performed by the manufacturer, guaranteeing the quality and specificity of the amplification. Non-specific signals (primer-dimers, amplification from non-specific primers), according to the manufacturer's instructions, appeared 8–10 cycles later than specific ones.
Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics 23, SNPstats and Arlequin 3.5 using the standard methods of genetic and statistical analysis (calculation of the frequency of alleles, individual genotypes, and their combinations). The strength of the association of genes, genotypes, and their combinations with UL was assessed using the odds ratio (OR) and the calculation of the 95% confidence interval. To assess the prognostic value of individual genotypes and their combinations, bioinformatic indicators such as specificity (Sp), positive predictive value of a test result, probability of developing a disease in an individual with a biomarker (PPV), negative predictive value of a test result, or probability of the absence of a disease in an individual without a biomarker (NPV) were calculated [12]. Data on patient age are presented as mean (M) and standard deviation (SD). The significance of differences in the frequencies of genetic traits in the study groups was determined using the two-tailed Fisher's exact test with Bonferroni adjustment for multiple comparisons calculated using the one-step method [13]. Differences were considered statistically significant at p<0.05. Genotype frequencies in the group of healthy women and patients were confirmed to be in Hardy–Weinberg equilibrium.
Results
Analysis of the distribution and structural characteristics of individual genotypes and multilocus combinations of 10 polymorphic positions of proinflammatory and anti-inflammatory cytokine genes included 2402 complex genotypes represented by combinations of 2–8 cytokine gene polymorphisms, 587 of which were statistically significant after Bonferroni adjustment for multiple comparisons.
Some of the multilocus combinations with the greatest strength of association with UL are presented in the table.
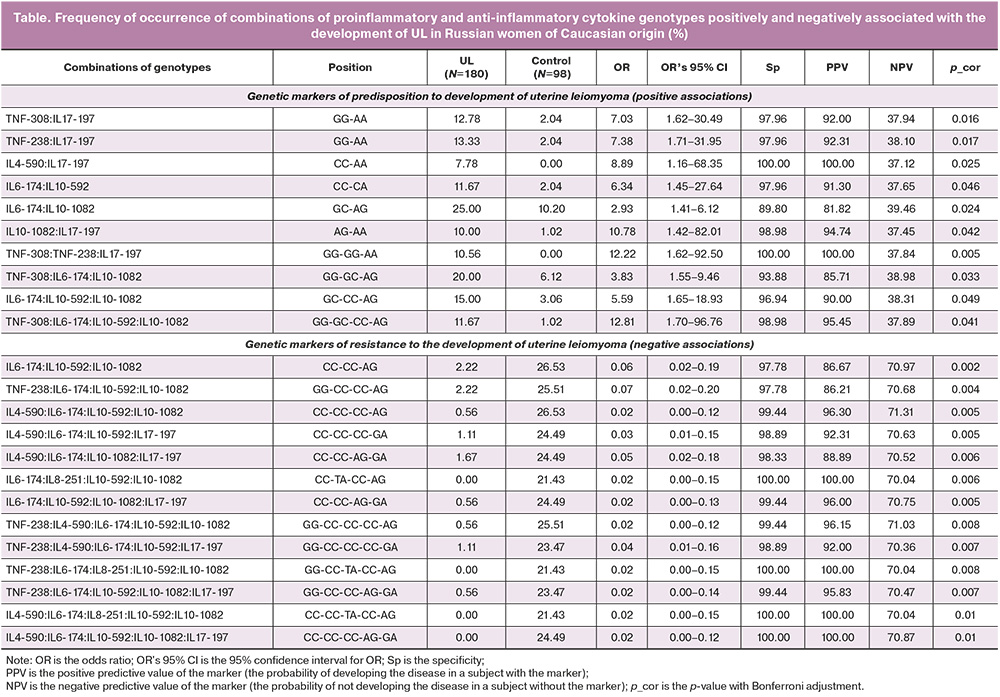
When comparing the frequencies of the combined genetic traits in the study groups with or without UL, two groups of indicators were identified. One of these groups was predominantly distributed among patients with UL (positive associations with the disease in 10 out of 587 biomarkers), while the other group was distributed only among women without clinical and instrumental signs of UL (negative associations with the disease in 577 biomarkers).
The dominant sequences in combinations that formed a predisposition to the development of UL were two- and 4-locus highly specific genetic patterns (10 gene chains of different sizes): [TNF-308 (IL17-197 in positions GG-AA] (OR=7.03, Sp=97.96%, p_cor=0.0162), [IL10-1082; IL17-197 in positions AG-AA] (OR=10.78, Sp=98.98%, p_cor=0.0423), and [TNF-308; IL6-174; L10-592; IL10-1082 in GG-GC-CC-AG] (OR=12.81, Sp=98.98%, p_cor=0.0414). In patients with UL, the genetic complexes differed from the control group (Table), and those genetic complexes predominated, whose frequency was significantly reduced in patients with UL compared to the control group. This suggests that in healthy women, combinations of cytokine gene SNPs with a protective effect, that is, those associated with resistance to UL development, predominate. Among this characteristics that can be conditionally classified as "resistant,” the number of SNPs included in their composition is somewhat wider than in the group of "predisposition" characteristics. Among them, SNPs of chemokine IL-8 and polymorphisms of cytokines with anti-inflammatory activity, IL-4 and IL-10 [IL6-174, IL8-251, IL10-592, IL10-1082 in CC-TA-CC-AG] (OR=0.02, Sp=100.00%, p_cor=0.0062) were more common; [IL4-590, IL6-174, IL8-251, IL10-592, and IL10-1082 in CC-CC-TA-CC-AG] (OR=0.02, Sp=100.00%, p_cor=0.0107). Considering the large number of resistance biomarkers detected in the control group, we can make a preliminary conclusion about the absence of polymorphisms as genetic risk factors for the development of UL in carriers of these compositions.
Sorting the identified complex genetic traits by the strength of their positive and negative associations with UL identified a group of highly specific complex genetic biomarkers with a positive predictive value (PPV) of up to 100%. For example, combinations of two variants of the IL4-590, IL17-197 genes in positions CC-AA or three variants of the TNF-308, TNF-238, IL17-197 in positions GG-GG-AA are not only characteristic of patients with UL but were also not detected in any patient in the control group (Sp=100.0%, p_cor< 0.05), which indicates a high prognostic value of these biomarkers in relation to the development of UL in a particular patient.
In contrast, a group of highly specific complex genetic traits (Sp>98%) was identified, the frequency of which was significantly lower in patients with UL than in the control group. Detection of such traits in a particular woman allows her to be classified as conditionally resistant to the development of UL with a probability (PPV) of 87–100%. In the same group of “resistant” genetic complexes, the negative predictive value (NPV) of a test result was also relatively high, that is, the probability of the absence of the disease in a subject without a marker reached 71.31% (p_cor=0.0056).
It is important to note that the relatively low NPV in most cases is due to the low frequency of the occurrence of complex biomarkers in the population. However, this general shortcoming of complex genetic markers can be compensated for by a broader set of markers in the prognostic matrix because of the search for new genetic compositions pathogenetically associated with the development of UL.
Discussion
In our study, we analyzed the results of an investigation of 10 SNPs in the cytokine genes. Although these 10 SNPs do not cover the entire cytokine genome, they include the most studied cytokine genes with proinflammatory and anti-inflammatory activities. Of particular interest in our data analysis was the wide representation of TNF gene variants in many SNP combinations. The cytokine encoded by this gene plays a crucial role in inflammation, apoptosis, immune system disorders, and tumor growth. Evidence suggests that the TC genotype and C allele at position -1031T/C (rs1799964) are associated with a predisposition to UL. The C allele -1031T/C promotes increased transcription, expression, and production of TNF-α [14], which can induce myocyte proliferation and the development of myomatous nodes. Furthermore, some of the cytokines we studied also have proangiogenic activity, which is important in the formation of myomatous nodes and requires the growth of the blood supply and activation of neoangiogenesis and neovasculogenesis. IL-6, for example, stimulates neovasculogenesis [15], while IL-8 (chemokine CXCL) is capable of stimulating angiogenesis by inducing the migration and proliferation of CXCR2-expressing endothelial cells, which ensures neovasculogenesis [16].
To date, several groups of genes associated with predisposition to the development of UL have been described. These include variants of genes, such as TP53, p21, XRCC1, VDR, transcription factor genes (MED12), growth factor genes (vascular endothelial growth factor gene (VEGF), IGF, and TGF-β), steroid receptor genes (estrogens and progesterone), inflammation-related genes (interleukins and TNF), and extracellular matrix remodeling genes (MMP and TIMP). Our results align with those of a study on a group of Russian women from the European part of the Russian Federation, which highlighted the significant role of molecular genetic markers of IL-10 (rs1800872), IL-5 (rs2069812), IL-4 (rs2243250), IL-1B (rs16944), and IL-1A (rs1800587) in UL development. These findings also emphasize the importance of interlocus interactions in determining predisposition to UL [20].
Our study focused on Russian women of European origin residing in the Siberian region of the Russian Federation. This is important due to the association between the prevalence of UL and the distribution of cytokine gene frequencies and polymorphic variants in relation to geographic and ethnic factors [21, 22]. While these associations complicate the extrapolation of our findings to the entire patient population, they underscore the need to develop personalized strategies for prognosis, early diagnosis, and treatment based on genetic factors [23].
Conclusion
A comparative analysis of the structure of genetic composites revealed that the specific compositions of pro- and anti-inflammatory cytokine genes are more significant in the pathogenesis of UL development than the individual polymorphisms. These compositions affect extracellular matrix remodeling, fibrosis, chronic inflammation, and oncogenesis. In our opinion, it is crucial to continue accumulating data and conducting bioinformatic analysis to identify highly informative genetic indicators of predisposition and resistance to UL. This will enable the implementation of targeted preventive measures and personalized approaches for a significant proportion of patients affected by this socially significant polygenic disease.
References
- Адамян Л.В., Кузнецова М.В., Тоноян Н.М., Шаповаленко Р.А., Пивазян Л.Г., Трофимов Д.Ю. Генетические аспекты миомы матки: современный взгляд на проблему. Проблемы репродукции. 2023; 29(4-2): 29-39. [Adamyan L.V., Kuznetsova M.V., Tonoyan N.M., Shapovalenko R.A., Pivazyan L.G., Trofimov D.Yu. Genetic aspects of uterine fibroids: a modern view of the problem. Russian Journal of Human Reproduction. 2023; 29(4-2): 29-39. (in Russian)]. https://dx.doi.org/10.17116/repro20232904229.
- Министерство здравоохранения Российской Федерации. Клинические рекомендации. Миома матки. 2020. [Ministry of Health of the Russian Federation. Clinical guidelines. Uterine fibroids. 2020. (in Russian)].
- Fedotova M., Barysheva E., Bushueva O. Pathways of hypoxia-inducible factor (HIF) in the orchestration of uterine fibroids development. Life (Basel). 2023; 13(8): 1740. https://dx.doi.org/10.3390/life13081740.
- Ramaiyer M.S., Saad E., Kurt I., Borahay M.A. Genetic mechanisms driving uterine leiomyoma pathobiology, epidemiology, and treatment. Genes (Basel). 2024; 15(5): 558. https://dx.doi.org/10.3390/genes15050558.
- Согоян Н.С., Кузнецова М.В., Донников А.Е., Мишина Н.Д., Михайловская Г.В., Шубина Е.С., Зеленский Д.В., Муллабаева С.М., Адамян Л.В. Семейная предрасположенность к развитию лейомиомы матки: поиск генетических факторов, повышающих риск развития заболевания. Проблемы репродукции. 2020; 26(5): 51-7. [Sogoyan N.S., Kuznetsova M.V., Donnikov A.E., Mishina N.D., Mikhailovskaya G.V., Shubina E.S., Zelenskii D.V., Mullabaeva S.M., Adamyan L.V. Familial predisposition to uterine leiomyoma: searching for genetic factors that increase the risk of leyomyoma development. Russian Journal of Human Reproduction. 2020; 26(5): 51-7. (in Russian)]. https://dx.doi.org/10.17116/repro20202605151.
- Кудрявцева О.К., Барышева Е.М., Вдовина И.Н., Клиновская А.А., Новикова Е.А., Полоников А.В., Иванов В.П., Бушуева О.Ю. Ассоциация полиморфных вариантов генов, вовлеченных в метаболизм глутатиона, с риском развития миомы матки. Медицинская генетика. 2020; 19(6): 52-4. [Kudryavtseva O.K., Barysheva E.M., Vdovina I.N., Klinovskaya A.A., Novikova E.A., Polonikov A.V., Ivanov V.P., Bushueva O.Yu. Association of genetic variations in genes involved in glutathione metabolism with risk of development of uterine fibroids. Medical Genetics. 2020; 19(6): 52-4. (in Russian)].
- Saad E.E., Michel R., Borahay M.A. Immunosuppressive tumor microenvironment and uterine fibroids: role in collagen synthesis. Cytokine Growth Factor Rev. 2024; 75: 93-100. https://dx.doi.org/10.1016/j.cytogfr.2023.10.002.
- Zhou X., Li Z., Zhou J. Tumor necrosis factor α in the onset and progression of leukemia. Exp. Hematol. 2017; 45: 17-26. https://dx.doi.org/10.1016/j.exphem.2016.10.005.
- Medikare V., Altaf A., Ananthapur V., Deendayal M., Nallari P. Susceptibility risk alleles of -238G/A, -308G/A and -1031T/C promoter polymorphisms of TNF-α gene to uterine leiomyomas. Recent Adv. DNA Gene Seq. 2015; 9(1): 65-71. https://dx.doi.org/10.2174/2352092210999151214155858.
- El-Tahan R.R., Ghoneim A.M., El-Mashad N. TNF-alpha gene polymorphisms and expression. Springerplus. 2016; 5(1): 1508. https://dx.doi.org/10.1186/s40064-016-3197-y.
- Хашукоева А.З., Хлынова С.А., Маркова Э.А., Каранашева А.Х. Миома матки: aлгоритм ведения и лечения. Акушерство и гинекология. 2021; 3(Приложение): 24-7. [Khashukoeva A.Z., Khlynova S.A., Markova E.A., Karanasheva A.Kh. Uterine myoma: algorithm for management and treatment. Obstetrics and Gynecology. 2021; (3 Suppl.): 24-7. (in Russian)].
- Simon R. Sensitivity, specificity, PPV, and NPV for predictive biomarkers. J. Natl. Cancer Inst. 2015; 107(8): djv153. https://dx.doi.org/10.1093/jnci/djv153.
- Наркевич А.Н., Виноградов К.А., Гржибовский А.М. Множественные сравнения в биомедицинских исследованиях: проблема и способы решения. Экология человека. 2020; 10: 55-64. [Narkevich A.N., Vinogradov K.A., Grzhibovskii A.M. Multiple comparisons in biomedical research: the problem and its solutions. Human Ecology. 2020; (10): 55-64. (in Russian)]. https://dx.doi.org/10.33396/1728-0869-2020-10-55-64.
- Nourian M., Chaleshi V., Pishkar L., Azimzadeh P., Baradaran Ghavami S., Balaii H. et al. Evaluation of tumor necrosis factor (TNF)-α mRNA expression level and the rs1799964 polymorphism of the TNF-α gene in peripheral mononuclear cells of patients with inflammatory bowel diseases. Biomed. Rep. 2017; 6(6): 698-702. https://dx.doi.org/10.3892/br.2017.908.
- Keshavarzi F., Salimi S., Mohammadpour-Gharehbagh A., Teimoori B., Yazdi A., Farajian-Mashhadi F. et al. The -2549 insertion/deletion polymorphism of VEGF gene associated with uterine leiomyoma susceptibility in women from Southeastern Iran. Ginekol. Pol. 2017; 88(3): 115-9. https://dx.doi.org/10.5603/GP.a2017.0022.
- Nenu I., Toadere T.M., Topor I., Țichindeleanu A., Bondor D.A., Trella S.E. et al. Interleukin-6 in hepatocellular carcinoma: a dualistic point of view. Biomedicines. 2023; 11(10): 2623. https://dx.doi.org/10.3390/biomedicines11102623.
- Upadhyay S., Dubey P.K. Gene variants polymorphisms and uterine leiomyoma: an updated review. Front. Genet. 2024; 15: 1330807. https://dx.doi.org/10.3389/fgene.2024.1330807.
- Королева Е.Г., Коненков В.И., Шевченко А.В., Прокофьев В.Ф., Орлов Н.Б., Тимофеева Ю.С., Айдагулова С.В., Маринкин И.О. Ассоциированность полиморфных вариантов генов цитокинов, фактора роста эндотелия и матричных металлопротеиназ с развитием миомы матки среди русских женщин. Сибирский научный медицинский журнал. 2024; 44(2): 113-22. [Koroleva Е.G., Konenkov V.I., Shevchenko A.V., Prokof’ev V.F., Orlov N.В., Timofeeva Yu.S., Aidagulova S.V., Marinkin I.О. Association of polymorphic variants of cytokines genes, endothelial growth factor and matrix metalloproteinases with the development of uterine fibroids among Russian women. Siberian Scientific Medical Journal. 2024; 44(2): 113-22. (in Russian)]. https://dx.doi.org/10.18699/SSMJ20240214.
- Maltese G., Fontanella C., Lepori S., Scaffa C., Fucà G., Bogani G. et al. Atypical uterine smooth muscle tumors: a retrospective evaluation of clinical and pathologic features. Oncology. 2018; 94(1): 1-6. https://dx.doi.org/10.1159/000479818.
- Алтухова О.Б., Радзинский В.Е., Сиротина С.С., Чурносов М.И., Ефремова О.А., Батлуцкая И.В., Орлова В.С. Полиморфизм генов интерлейкинов и риск развития миомы матки. Акушерство и гинекология. 2022; 7: 81-7. [Altukhova O.B., Radzinskii V.E., Sirotina S.S., Churnosov M.I., Efremova O.A., Batlutskaya I.V., Orlova V.S. Interleukin gene polymorphism and risk of uterine fibroids. Obstetrics and Gynecology. 2022; (7): 81-7. (in Russian)]. https://dx.doi.org/10.18565/aig.2022.7.81-87.
- Yu O., Scholes D., Schulze-Rath R., Grafton J., Hansen K., Reed S.D. A US population-based study of uterine fibroid diagnosis incidence, trends, and prevalence: 2005 through 2014. Am. J. Obstet. Gynecol. 2018; 219(6): 591.e1-591.e8. https://dx.doi.org/10.1016/j.ajog.2018.09.039.
- Laughlin-Tommaso S.K., Jacoby V.L., Myers E.R. Disparities in fibroid incidence, prognosis, and management. Obstet. Gynecol. Clin. North Am. 2017; 44(1): 81-94. https://dx.doi.org/10.1016/j.ogc.2016.11.007.
- Koltsova A.S., Efimova O.A., Pendina A.A. A view on uterine leiomyoma genesis through the prism of genetic, epigenetic and cellular heterogeneity. Int. J. Mol. Sci. 2023; 24(6): 5752. https://dx.doi.org/10.3390/ijms 24065752.
Received 03.07.2024
Accepted 19.08.2024
About the Authors
Vladimir I. Konenkov, Dr. Med. Sci., Professor, Academician of the RAS, Head of the Clinical Immunogenetics Laboratory, Research Institute of Clinical and Experimental Lymрhology – Branch of the Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences, 2 Timakova str., Novosibirsk, 630060, Russia,+7(383)333-64-09, vikonenkov@gmail.com, https://orcid.org/0000-0001-7385-6270
Elena G. Koroleva, obstetrician-gynecologist, Junior Researcher at the Laboratory of Cellular Biology and Fundamental Basis of Reproduction of Central Scientific Laboratory, Novosibirsk State Medical University, Ministry of Health of Russia, 52 Krasnyy prospect str., Novosibirsk, 630091, Russia, +7(383)226-35-60, korlex71@mail.ru,
https://orcid.org/0000-0002-8522-4382
Viktor F. Prokofiev, PhD, Leading Researcher at Clinical Immunogenetics Laboratory, Research Institute of Clinical and Experimental Lymрhology – Branch of the Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences, 13 Frunze str./15 Michurina str., Novosibirsk, 630091, Russia, +7(383)373-96-01, vf_prok@mail.ru, https://orcid.org/0000-0001-7290-1631
Alla V. Shevchenko, Dr. Biol. Sci., Leading Researcher at Clinical Immunogenetics Laboratory, Research Institute of Clinical and Experimental Lymрhology – Branch of the Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences, 13 Frunze str./15 Michurina str., Novosibirsk, 630091, Russia, +7(383)373-96-01,
https://orcid.org/0000-0001-5898-950X
Yulia S. Timofeeva, PhD, Teaching Assistant at the Obstetrics and Gynecology Department, Novosibirsk State Medical University, Ministry of Health of Russia,
52 Krasnyy prospect str., Novosibirsk, 630091, Russia, +7(961)220-31-13, dr.j.timofeeva@yandex.ru, https://orcid.org/0000-0002-5379-9296
Svetlana V. Aidagulova, Dr. Biol. Sci., Professor, Head of the Laboratory of Cellular Biology and Fundamental Basis of Reproduction of Central Scientific Laboratory, Novosibirsk State Medical University, Ministry of Health of Russia, 52 Krasnyy prospect str., Novosibirsk, 630091, Russia, +7(913)909-22-51, asvetvlad@yandex.ru,
https://orcid.org/0000-0001-7124-1969
Igor O. Marinkin, Dr. Med. Sci., Professor, Head of the Obstetrics and Gynecology Department, Rector, Novosibirsk State Medical University, Ministry of Health of Russia,
52 Krasnyy prospect str., Novosibirsk, 630091, Russia, +7(383)222-32-04, rector@ngmu.ru, https://orcid.org/0000-0002-9409-4823
Corresponding author: Viktor F. Prokofiev, vf_prok@mail.ru



