Association between xenobiotic detoxification system gene polymorphisms and hormone replacement therapy efficacy in women with premature ovarian insufficiency
Averkova V.G., Donnikov A.E., Yureneva S.V.
Objective: To evaluate the association between xenobiotic detoxification system gene polymorphisms and hormone replacement therapy (HRT) efficacy in patients with premature ovarian insufficiency (POI).
Materials and methods: This study included 83 women with POI who exhibited persistent symptoms of estrogen deficiency while receiving HRT E2/DYD 2 mg/10 mg. The participants were divided into two groups: group 1 (n=23) included patients with signs of severe estrogen deficiency (GCS score > 20 and E2 blood level <150 pmol/l) and group 2 (n=60) included those with a GCS score < 20 and an E2 blood level >150 pmol/l. Genotyping was performed by real-time PCR. The distribution frequencies of alleles and genotypes for the polymorphisms were analyzed in both groups. The χ² test assessed the significance of the differences (p). The strength of the association between features was evaluated using the odds ratio (OR).
Results: Allele A of the A313G polymorphism of GSTP1 and allele C of the C341T polymorphism of GSTP1 were associated with severe estrogen deficiency in patients receiving HRT E2/DYD 2 mg/10 mg (OR=2.99, 95% CI 1.07–8.33, p=0.03; OR=7.43, 95% CI 0.96–57.52, p=0.03). The AC haplotype (risk haplotype) for the GSTP1 A313G (Ile105Val) and GSTP1 C341T (Ala114Val) polymorphisms was significantly more frequently detected in patients with severe estrogen deficiency during HRT (69.6% in group 1 versus 43.3% in group 2, p=0.049, OR=2.99, 95% CI 0.97–9.80). The -341 C/C genotype was identified as a marker of the risk haplotype based on haplotype and individual genotype correspondence assessment.
Conclusion: GSTP1 gene polymorphisms play a significant role in the response to HRT in patients with POI, likely due to their influence on the regulation of E2 detoxification processes.
Authors' contributions: Averkova V.G. – obtaining data for analysis, material processing and analyzing, statistical analysis, drafting of the manuscript; Donnikov A.E. – editing of the manuscript; Yureneva S.V. – conception and design of the study, editing of the manuscript.
Conflicts of interest: The authors have no conflicts of interest to declare.
Funding: There was no funding for this study.
Ethical Approval: The study was reviewed and approved by the Research Ethics Committee of the V.I. Kulakov NMRC for OG&P (Ref. No: 1 of 07 February 2019).
Patient Consent for Publication: All patients provided informed consent for the publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available upon request from the corresponding author after approval from the principal investigator.
For citation: Averkova V.G., Donnikov A.E., Yureneva S.V. Association between xenobiotic detoxification system gene polymorphisms and hormone replacement therapy efficacy in women with premature ovarian insufficiency.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2024; (10): 113-120 (in Russian)
https://dx.doi.org/10.18565/aig.2024.243
Keywords
Premature ovarian insufficiency (POI) is defined as the loss of ovarian function in women under 40 years of age [1]. According to a 2023 systematic review and meta-analysis, the global prevalence of POI has been increasing over the past 20 years and now represents 3.5% of the female population [2]. The primary strategy for treating women with this disorder is long-term hormone replacement therapy (HRT) with sex steroids until the average age of natural menopause, thereby mimicking normal ovarian function [3]. Approaches to prescribing specific doses and compositions of HRT for POI are currently under discussion. However, major guidelines indicate that patients with POI usually require higher doses of hormones (especially estrogens) than those prescribed for women experiencing timely menopause [1–4]. Our previous study showed that the most commonly used HRT in women with POI was 2 mg oral estradiol (E2) in the form of E2/dydrogesterone (DYD) 2 mg/10 mg [5]. Nevertheless, the efficacy of this therapy and patient satisfaction with the treatment vary widely. Some patients still exhibit signs of severe estrogen deficiency despite regular HRT. It is well known that the efficacy and safety of drug therapy are influenced by various clinical and demographic factors such as gender, ethnicity, age, weight, and drug interactions [6]. Furthermore, the response to treatment depends on the genetic characteristics of individuals, particularly the presence of variants in the genes encoding enzymes involved in the biotransformation of medicinal agents (MA). The activity of the biotransformation system is a key factor that determines the pharmacokinetics of MA, and consequently, the patient's pharmacological response to therapy [6, 7]. Studies have shown that four out of five patients are likely to be carriers of genetic variants that can alter the effectiveness of the prescribed MA [7]. Pharmacogenetic testing (PGT) is of clinical importance. By utilizing molecular genetic markers, it is possible to identify individual patient characteristics that determine their response to prescribed therapies [8]. Currently, PGT has enabled the development of personalized algorithms for prescribing MA for various pathologies [9, 10].
Pharmacogenetics of menopause and primary ovarian insufficiency (POI) are still in the early stages of development. Hormone replacement therapy (HRT) drugs, such as menopausal hormone therapy (MHT), are metabolized according to the same principles as other medications and are converted into active compounds that provide a therapeutic effect through the action of phase I and II enzymes of the xenobiotic detoxification system, and subsequently eliminated from the body as metabolites [11]. Research on gene variants responsible for the detoxification of exogenous estrogens has primarily focused on their role in the development of adverse events associated with MHT in postmenopausal women, including breast cancer [12, 13].
Studies examining individual differences in response to MHT are even more limited. The effectiveness of MHT in postmenopausal patients, influenced by variants in genes associated with estrogen metabolism, was explored in a large-scale study called the Kronos Early Estrogen Prevention Study (KEEPS). The results of this study indicated that specific single nucleotide polymorphisms (SNPs) in the SULT1A1 gene, which encodes the phase II enzyme sulfotransferase 1A1 (SULT1A1) involved in exogenous estrogen detoxification, are associated with an earlier age of menopause and greater effectiveness of HRT in addressing sleep disorders and night sweats [14]. However, no previous studies have investigated the impact of gene variants in the detoxification system on the effectiveness of HRT in women with POI.
This study aimed to evaluate the effect of polymorphisms in genes of the xenobiotic detoxification system (GSTP1 A313G, GSTP1 C341T, GPX1 C599T, EPHX1 T337C, EPHX1 A416G, SOD2 T47C, NAT2 A590G, NAT2 A857G, SULT1A1 A638G, CYP1A1 A1506G, and CYP1A1 T6235C) on the effectiveness of HRT in patients with POI.
Materials and methods
The study was conducted at the V.I. Kulakov NMRC for OG&P of the Ministry of Health of Russia and reviewed and approved by the local Research Ethics Committee. The study included 83 women with primary ovarian insufficiency (POI) who had persistent symptoms of estrogen deficiency while receiving HRT E2/DYD 2 mg/10 mg. All patients signed an informed consent form to participate in this study.
Patients were selected based on predefined inclusion and exclusion criteria.
The inclusion criteria for the study were a normal karyotype, spontaneous POI, HRT intake duration of at least 6 months, and age of ≥18 and <45 years.
Exclusion criteria included iatrogenic POI, severe somatic comorbidities, contraindications to HRT (according to the clinical guidelines of the Ministry of Health of the Russian Federation "Menopause and Climacteric State in Women" 2021 and drug instructions), thyroid pathology with decompensation of function, diagnosed mental disorders, current psychopharmacotherapy, and the use of dietary supplements containing St. John's Wort.
POI diagnosis was established according to the diagnostic criteria adopted in 2016 by the European Society of Human Reproduction and Embryology (ESHRE). These criteria included oligo/amenorrhea for at least 4 months, follicle-stimulating hormone (FSH) levels > 25 mIU/L confirmed twice at an interval of 4–6 weeks, and age of women being under 40 years. The severity of estrogen deficiency symptoms was assessed using the Greene Climacteric Scale (GCS) questionnaire. A blood test for E2 was conducted using enzyme immunoassay on the 2nd or 3rd day of the induced menstrual cycle, 23–24 h after taking the next E2/DYD 2 mg/10 mg tablet.
After the examination and evaluation of the effectiveness of HRT, the study participants were divided into two groups. Group 1 (n=23) included patients who, despite HRT E2/DYD 2 mg/10 mg, still exhibited signs of severe estrogen deficiency (more than 20 points on the GCS questionnaire and E2 levels in the blood below the laboratory's lower reference limit of 150 pmol/L). Group 2 included all other patients (n=60).
Molecular genetic studies were conducted at the Laboratory of Molecular Genetics of V. I. Kulakov NMRC for OG&P of the Ministry of Health of Russia. Genomic DNA, isolated from peripheral venous blood collected in EDTA (ethylenediaminetetraacetic acid) as an anticoagulant, served as the material for the study. The Proba-GS-Genetics reagent kit (NPO DNA-Technology, LLC, Russia) was used for isolation. Polymerase chain reaction and determination of the melting temperature of the oligonucleotide probes were performed using a DT-964 detection amplifier (NPO DNA Technology, Russia).
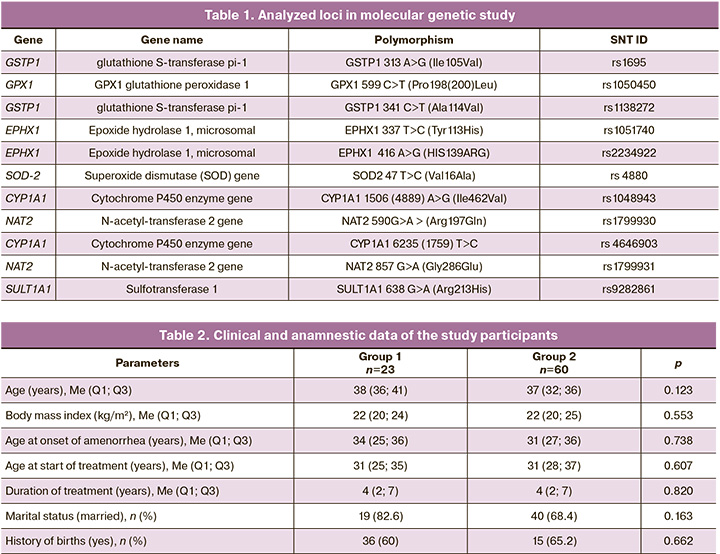
The loci analyzed are listed in Table 1.
Statistical analysis
Statistical analysis was performed using SPSS (IBM Statistical Package for the Social Sciences, version 25). Descriptive statistics are presented as counts with percentages for categorical variables, and median (Me) and interquartile range (Q1; Q3) for continuous variables. In the analysis of data from molecular genetic studies, the χ2 test was used to determine the significance of differences in the frequency of the occurrence of qualitative characteristics. When assessing the association of an allele with a phenotypic characteristic, the genotypic frequencies of the analyzed allele were compared between the groups of patients with and without this characteristic. The distribution of genotypes in the groups was compared based on the presence of significant differences in the distribution of alleles. In this case, the hypotheses of autosomal dominant and autosomal recessive inheritance of the analyzed characteristics were tested by constructing 2×2 contingency tables. For each table, Fisher's exact test (F) was used if the expected frequency of one or more cells was less than five, or the χ2 test. A linkage analysis was performed for these polymorphisms. The odds ratio (OR) for the manifestation of the characteristic with the corresponding genotype was also calculated. The critical significance level for testing the statistical hypotheses was set at p<0.05.
Results
The clinical, anamnestic, and anthropometric data of the patients in groups 1 and 2 did not differ (Table 2).
The distribution of genotypes was consistent with that expected under Hardy-Weinberg equilibrium for all genes studied. The distribution of alleles and genotypes at the polymorphic loci of the xenobiotic detoxification system genes, based on the effectiveness of HRT E2/DYD 2 mg/10 mg in patients with POI, was as follows: the frequency of the A allele of the A313G polymorphism of the GSTP1 gene in group 1 (83%) was significantly higher than that in group 2 (65%) (OR=2.56, 95% CI 1.09–5.98, p=0.03). According to the autosomal recessive model of inheritance, the A allele of the A313G polymorphism of GSTP1 was associated with low efficiency of HRT and signs of severe estrogen deficiency (OR=2.99, 95% CI 1.07–8.33, p=0.03). The frequency of the C allele of the C341T polymorphism of the GSTP1 gene in group 1 was also statistically significantly higher compared to group 2 (98% versus 86%, respectively), with an OR of 7.43 (95% CI 0.96–57.52, p=0.03). There were differences between the two groups of patients regarding the alleles of the GPX1 C599T polymorphism; however, these differences did not reach statistical significance: 78% in group 1 versus 63% in group 2 (OR=2.08, 95% CI 0.94–4.61, p=0.07). At the same time, according to the autosomal recessive model of inheritance, the C/C genotype of the C599T polymorphism of the GPX1 gene was associated with low HRT efficiency and signs of severe estrogen deficiency in patients (OR=3.02, 95% CI 1.11–8.23, p=0.03). It should be noted that some of the studied polymorphisms of GSTP1 are located close to the chromosome (890 nucleotide sequences), suggesting the possibility of linked inheritance of certain alleles. Linkage analysis of the polymorphisms revealed linked inheritance of the polymorphic loci Ile105Val and Ala114Val: D’=1.0 (0.75–1.0), LOD=7.24. A search for associations between HRT efficiency indicators and the carriage of haplotypes for the polymorphic loci of GSTP1, Ile105Val and Ala114Val, was conducted. Table 3 presents the incidence of haplotypes for polymorphisms of the selected genes in the study groups.
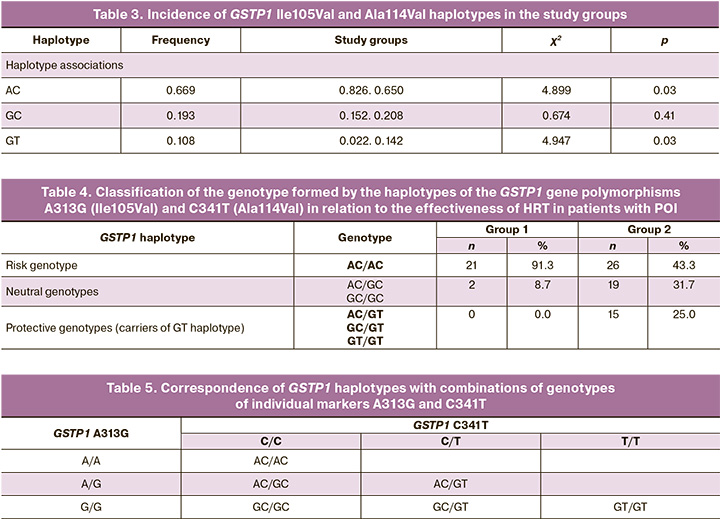
Haplotype AC was more frequently detected in patients of group 1 (69.6%) compared to group 2 (43.3%; p=0.03, OR=2.99, 95% CI 0.97–9.80), while haplotype GT was more common in patients of group 2 (3.3%) compared to group 1 (0.0%; p=0.03, OR=7.33, 95% CI 0.98–322.5). Thus, haplotype AC can be considered a risk factor, whereas haplotype GT serves as a protective factor against the low efficiency of HRT E2/DYD 2 mg/10 mg. The frequency of occurrence of the GC haplotype did not differ significantly between the studied patient groups; therefore, it can be characterized as a neutral factor.
Clinical and functional assessment of the genotype of the GSTP1 gene polymorphisms A313G (Ile105Val) and C341T (Ala114Val) revealed that the combination of two protective haplotypes, protective and neutral, or a protective haplotype with a risk haplotype (i.e., carriage of the GT haplotype), was regarded as a protective genotype (OR=2.1, 95% CI 1.54–2.89, p<0.0001). The combination of the two risk alleles was classified as a risk genotype (OR=0.0, 95% CI 0.0–0.6, p=0.008). The combination of a risk allele with a neutral allele as well as a combination of two neutral alleles was considered a neutral genotype (Table 4).
The standard approach in routine clinical practice is to determine single nucleotides at functionally significant haplotype marker points rather than the haplotypes themselves, due to the technological complexity and high cost of such studies. The complete genotype was established based on data regarding linkage during inheritance of the determined marker points. The correspondence of haplotypes and individual genotypes of GSTP1 A313G (Ile105Val) and C341T (Ala114Val) is presented in Table 5.
The marker of the protective haplotype was the genotype -313 G/G, whereas the marker of the risk haplotype was the genotype -341 C/C. The combination of -313 G/G and -341 C/C, as well as the two heterozygous genotypes, served as a marker of the neutral haplotype.
Discussion
According to the International Menopause Society (IMS) in their statement "Pharmacogenomics in personalized medicine: menopause perspectives," a personalized approach to treating climacteric syndrome and menopause-related conditions should incorporate pharmacogenomics [16]. This approach helps optimize therapy for estrogen deficiency, as there are significant inter-individual differences in the effective dose of estradiol (E2) required in hormone replacement therapy (HRT) to alleviate symptoms. The effective doses of HRT needed to relieve estrogen deficiency symptoms and prevent age-associated diseases (e.g., osteoporosis and cardiovascular diseases) may differ between young women with premature ovarian insufficiency (POI) or early menopause and those experiencing timely menopause [16].
In this study, we conducted a genetic analysis of variants in genes involved in phase I and II xenobiotic detoxification, and examined their association with the effectiveness of HRT (E2/DYD 2 mg/10 mg) in patients with POI. The response to therapy was associated with variants in the GSTP1 gene, which encodes the phase II enzyme glutathione-S-transferase P1, which is responsible for xenobiotic detoxification. This enzyme is widely expressed throughout the body, except in erythrocytes [17]. GSTP1 is located on the long arm of chromosome 11 (11q13) and has two well-studied polymorphisms involving single-nucleotide substitutions in the coding regions (1578A>G and 2293C>T): an adenine-to-guanine (A/G) substitution in exon 5 (position 313), and a cytosine-to-thymine (C/T) substitution in exon 6 (position 341). These amino acid substitutions occur in the enzyme's active site, leading to differences in the enzymatic activity of the protein product depending on the allele [18].
Several studies have examined the frequency of GSTP1 polymorphisms in different populations. The GSTP1 114Val variant occurs in approximately 10% of cases, and the GSTP1 105Val variant occurs in 33% of Caucasians [19]. The frequency of homozygosity for the wild-type allele GSTP1 105Val in Europeans is approximately 40–60% [20]. According to literature, glutathione-S-transferase P1 exhibits the highest activity in individuals homozygous for the wild-type allele (A) (Ile/Ile), reduced activity in heterozygous carriers (Ile/Val), and the lowest activity in homozygous carriers of the mutant allele (Val/Val) [21].
In our study, both variants of GSTP1 were associated with the response to HRT (E2/DYD 2 mg/10 mg). Linkage analysis revealed statistically significant differences in the frequency of haplotypes for GSTP1 A313G (Ile105Val) and GSTP1 C341T (Ala114Val) polymorphisms. The AC haplotype was found more frequently in patients with severe estrogen deficiency, whereas the GT haplotype was found in patients without severe estrogen deficiency during HRT. To further support its clinical application, we identified markers for the risk haplotype (genotype -341 C/C) and protective haplotype (genotype -313 G/G) associated with low HRT effectiveness (E2/DYD 2 mg/10 mg).
We were unable to find studies that directly examined the association between GSTP1 variants and the effectiveness of HRT in patients with POI. Similar findings have also been reported in other studies. For instance, the GSTP1 A313G (Ile105Val) polymorphism is associated with a better response to cyclophosphamide treatment in autoimmune diseases. The authors hypothesized that reduced enzyme activity leads to decreased conjugation and elimination of cyclophosphamide, resulting in higher therapeutic drug concentrations [22, 23].
The role of GSTP1 polymorphisms in the efficacy of various chemotherapeutic agents has been well-studied in oncology. Ji M. et al. reported that the AA genotype at the GSTP1 A313G polymorphic locus was linked to a poorer response to neoadjuvant chemotherapy in breast cancer patients, attributed to increased drug conjugation and elimination [24]. In contrast, the A→G substitution of GSTP1 Ile105Val was associated with a better response to platinum-based chemotherapy in advanced non-small cell lung cancer [25, 26], melphalan treatment for multiple myeloma [27], and chemotherapy for colorectal cancer [28]. Additionally, a meta-analysis of 21 randomized controlled trials (RCTs) involving 4,990 participants confirmed that variants in other sulfotransferase genes, leading to reduced enzymatic activity, were associated with greater chemotherapy efficacy [29].
Notably, conflicting results regarding GSTP1 polymorphisms have been reported. For example, McIlwain C.C. et al. found that carriers of GSTP1 (A/G + G/G) allelic variants had a poorer response to tyrosine kinase inhibitors than homozygous wild-type allele carriers (A/A) [30]. These discrepancies may be due to substrate specificity, as variants in the GSTP1 gene likely impact enzyme activity differently, depending on the drug. For instance, the GSTP1 105Val variant decreased glutathione-S-transferase P1 catalytic activity two-fold for thiotepa and 15-fold for chlorambucil [31]. Ishimoto T.M. and Ali-Osman F., using in vitro models, demonstrated that the cytoprotective effect of GSTP1 variants varies significantly depending on the drug [32].
A recent study using GSTP1 knockout mice (Gstp1 / p2 -/-) provided further insights, showing an association between GSTP1 and the activity of the estrogen receptor (ERα). Glutathione-S-transferases formed a protein complex with ERα, reducing its binding potential to E2 by threefold [33]. Thus, GSTP1 polymorphisms may influence the response to HRT in patients with POI through multiple mechanisms. However, given the limited available data and complexity of drug metabolism, further research with larger sample sizes and additional genes is warranted.
Conclusion
The GSTP1 variants Ile105Val and Ala114Val play a significant role in shaping the response to HRT in patients with POI, likely through the regulation of E2 detoxification processes. Identification of specific haplotypes and genotypes (GSTP1 A313G and C341T) simplifies genetic testing in routine clinical practice. These findings provide a genomic basis for variability in therapy response, aiding in the development of personalized HRT regimens for patients with POI. Carriers of GSTP1 polymorphic variants may require higher doses of hormonal drugs owing to the reduced efficacy of standard E2 doses in HRT.
References
- Panay N., Anderson R., Bennie A., Cedars M., Davies M., Ee C. et al. O-111 Premature ovarian insufficiency: new data and updated guidance. Hum. Reprod. 2024; 39(Suppl._1): i62. https://dx.doi.org/10.1093/humrep/deae108.122.
- Li M., Zhu Y., Wei J., Chen L., Chen S., Lai D. The global prevalence of premature ovarian insufficiency: a systematic review and meta-analysis. Climacteric. 2023; 26(2): 95-102. https://dx.doi.org/10.1080/13697137.2022.2153033.
- Panay N., Anderson R.A., Nappi R.E., Vincent A.J., Vujovic S., Webber L. et al. Premature ovarian insufficiency: an International Menopause Society White Paper. Climacteric. 2020; 23(5): 426-46. https://dx.doi.org/10.1080/13697137.2020.1804547.
- Jayasena C.N., Devine K., Barber K., Comninos A.N., Conway G.S., Crown A. et al. Society for endocrinology guideline for understanding, diagnosing and treating female hypogonadism. Clin. Endocrinol. (Oxf.). 2024; 101(5): 409-42. https://dx.doi.org/10.1111/cen.15097.
- Аверкова В.Г., Юренева С.В. Анализ удовлетворенности лечением пациенток с преждевременной недостаточностью яичников. Акушерство и гинекология. 2022; 10: 83-92. [Averkova V.G., Yureneva S.V. Analysis of treatment satisfaction in patients with premature ovarian failure. Obstetrics and Gynecology. 2022; (10): 83-92. (in Russian)]. https://dx.doi.org/10.18565/aig.2022.10.83-92.
- Кукес В.Г., Сычев Д.А., Раменская Г.В., Игнатьев И.В. Фармакогенетика системы биотрансформации и транспортеров лекарственных средств: от теории к практике. Биомедицина. 2007; 6: 29-47. [Kukes V.G., Sychev D.A., Ramenskaya G.V., Ignat’ev I.V. Pharmacogenetics of system of biotransformation and drugs transporters: from the theory to practice. Biomedicine. 2007; (6): 29-47. (in Russian)].
- Chenchula S., Atal S., Uppugunduri C.R.S. A review of real-world evidence on preemptive pharmacogenomic testing for preventing adverse drug reactions: a reality for future health care. Pharmacogenomics J. 2024; 24(2): 9. https://dx.doi.org/10.1038/s41397-024-00326-1.
- Zhou Z.W., Chen X.W., Sneed K.B., Yang Y.X., Zhang X., He Z.X. et al. Clinical association between pharmacogenomics and adverse drug reactions. Drugs. 2015; 75(6): 589-631. https://dx.doi.org/10.1007/s40265-015-0375-0.
- Thomas C.D., Johnson J.A. Pharmacogenetic factors affecting β-blocker metabolism and response. Expert Opin. Drug Metab. Toxicol. 2020; 16(10): 953-64. https://dx.doi.org/10.1080/17425255.2020.1803279.
- Asiimwe I.G., Pirmohamed M. Ethnic diversity and warfarin pharmacogenomics. Front. Pharmacol. 2022; 13: 866058. https://dx.doi.org/10.3389/fphar.2022.866058.
- Almazroo O.A., Miah M.K., Venkataramanan R. Drug metabolism in the liver. Clin. Liver Dis. 2017; 21(1): 1-20. https://dx.doi.org/10.1016/j.cld.2016.08.001.
- Cerne J.Z., Novakovic S., Frkovic-Grazio S., Pohar-Perme M., Stegel V., Gersak K. Estrogen metabolism genotypes, use of long-term hormone replacement therapy and risk of postmenopausal breast cancer. Oncol. Rep. 2011; 26(2): 479-85. https://dx.doi.org/10.3892/or.2011.1298.
- MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk. Genetic polymorphisms in phase I and phase II enzymes and breast cancer risk associated with menopausal hormone therapy in postmenopausal women. Breast Cancer Res. Treat. 2010; 119(2): 463-74. https://dx.doi.org/10.1007/s10549-009-0407-0.
- Miller V.M., Naftolin F., Asthana S., Black D.M., Brinton E.A., Budoff M.J. et al. The Kronos Early Estrogen Prevention Study (KEEPS): what have we learned? Menopause. 2019; 26(9): 1071-84. https://dx.doi.org/10.1097/GME.0000000000001326.
- European Society for Human Reproduction and Embryology (ESHRE) Guideline Group on POI; Webber L., Davies M., Anderson R., Bartlett J., Braat D., Cartwright B. et al. ESHRE Guideline: management of women with premature ovarian insufficiency. Hum. Reprod. 2016; 31(5): 926-37. https://dx.doi.org/10.1093/humrep/dew027.
- Moyer A.M., Miller V.M., Faubion S.S. Could personalized management of menopause based on genomics become a reality? Pharmacogenomics. 2016; 17(7): 659-62. https://dx.doi.org/10.2217/pgs.16.17.
- Potęga A. Glutathione-mediated conjugation of anticancer drugs: an overview of reaction mechanisms and biological significance for drug detoxification and bioactivation. Molecules. 2022; 27(16): 5252. https://dx.doi.org/10.3390/molecules27165252.
- Gatedee J., Pakakassama S., Muangman S., Pongstaporn W. Glutathione S-transferase P1 genotypes, genetic susceptibility and outcome of therapy in thai childhood acute lymphoblastic leukemia. Asian Pac. J. Cancer Prev. 2007; 8(2): 294-6.
- Dasgupta R.K., Adamson P.J., Davies F.E., Rollinson S., Roddam P.L., Ashcroft A.J. et al. Polymorphic variation in GSTP1 modulates outcome following therapy for multiple myeloma. Blood. 2003; 102(7): 2345-50. https://dx.doi.org/10.1182/blood-2003-02-0444.
- Фетисова И.Н., Межинский С.С., Чаша Т.В., Ратникова С.Ю., Фетисов Н.С. Полиморфизм генов системы детоксикации. Вестник Ивановской медицинской академии. 2014; 19(4): 50-8. [Fetisova I.N., Mezhinskii S.S., Chasha T.V., Ratnikova S.Yu., Fetisov N.S. Gene polymorphism of detoxication system. Bulletin of Ivanovo State Medical Academy. 2014; 19(4): 50-8. (in Russian)].
- Garte S., Gaspari L., Alexandrie A.K., Ambrosone C., Autrup H., Autrup J.L. et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol. Biomarkers Prev. 2001; 10(12): 1239-48.
- Hajdinák P., Szabó M., Kiss E., Veress L., Wunderlich L., Szarka A. Genetic polymorphism of GSTP-1 affects cyclophosphamide treatment of autoimmune diseases. Molecules. 2020; 25(7): 1542. https://dx.doi.org/10.3390/molecules25071542.
- Attia D.H.S., Eissa M., Samy L.A., Khattab R.A. Influence of glutathione S transferase A1 gene polymorphism (-69C > T, rs3957356) on intravenous cyclophosphamide efficacy and side effects: a case-control study in Egyptian patients with lupus nephritis. Clin. Rheumatol. 2021; 40(2): 753-62. https://dx.doi.org/10.1007/s10067-020-05276-0.
- Ji M., Tang J., Zhao J., Xu B., Qin J., Lu J. Polymorphisms in genes involved in drug detoxification and clinical outcomes of anthracycline-based neoadjuvant chemotherapy in Chinese Han breast cancer patients. Cancer Biol. Ther. 2012; 13(5): 264-71. https://dx.doi.org/10.4161/cbt.18920.
- Sun N., Sun X., Chen B., Cheng H., Feng J., Cheng L. et al. MRP2 and GSTP1 polymorphisms and chemotherapy response in advanced non-small cell lung cancer. Cancer Chemother. Pharmacol. 2010; 65(3): 437-46. https://dx.doi.org/10.1007/s00280-009-1046-1.
- Lv H., Han T., Shi X., Yao Y., Yao Y., Qiu W. et al. Genetic polymorphism of GSTP1 and ERCC1 correlated with response to platinum-based chemotherapy in non-small cell lung cancer. Med. Oncol. 2014; 31(8): 86. https://dx.doi.org/10.1007/s12032-014-0086-5.
- Dasgupta R.K., Adamson P.J., Davies F.E., Rollinson S., Roddam P.L., Ashcroft A.J. et al. Polymorphic variation in GSTP1 modulates outcome following therapy for multiple myeloma. Blood. 2003; 102(7): 2345-50. https://dx.doi.org/10.1182/blood-2003-02-0444.
- Yiannakopoulou E.Ch. Pharmacogenomics of phase II metabolizing enzymes and drug transporters: clinical implications. Pharmacogenomics J. 2013; 13(2): 105-9. https://dx.doi.org/10.1038/tpj.2012.42.
- Hu X.Y., Huang X.Y., Ma J., Zuo Y., Luo N.B., Lai S.L. et al. GSTT1 and GSTM1 polymorphisms predict treatment outcome for breast cancer: a systematic review and meta-analysis. Tumour Biol. 2016; 37(1): 151-62. https://dx.doi.org/10.1007/s13277-015-4401-3.
- McIlwain C.C., Townsend D.M., Tew K.D. Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene. 2006; 25(11): 1639-48. https://dx.doi.org/10.1038/sj.onc.1209373.
- Pandya U., Srivastava S.K., Singhal S.S., Pal A., Awasthi S., Zimniak P. et al. Activity of allelic variants of Pi class human glutathione S-transferase toward chlorambucil. Biochem. Biophys. Res. Commun. 2000; 278(1): 258-62. https://dx.doi.org/10.1006/bbrc.2000.3787.
- Ishimoto T.M., Ali-Osman F. Allelic variants of the human glutathione S-transferase P1 gene confer differential cytoprotection against anticancer agents in Escherichia coli. Pharmacogenetics. 2002; 12(7): 543-53. https://dx.doi.org/10.1097/00008571-200210000-00006.
- Zhang J., Ye Z.W., Chen W., Manevich Y., Mehrotra S., Ball L. et al. S-Glutathionylation of estrogen receptor α affects dendritic cell function. J. Biol. Chem. 2018; 293(12): 4366-80. https://dx.doi.org/10.1074/jbc.M117.814327.
Received 07.10.2024
Accepted 17.10.2024
About the Authors
Victoria G. Averkova, Researcher, Institute of Oncology and Mammology, V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, 117997, Russia, Moscow, Oparin str., 4, buch1202@mail.ru, https://orcid.org/0000-0002-8584-5517Svetlana V. Yureneva, Dr. Med. Sci., Professor at the Department of Obstetrics and Gynecology of the Department of Vocational Education, Deputy Director for Science, Institute of Oncology and Mammology, V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, 117997, Russia, Moscow, Oparin str., 4, syureneva@gmail.com,
https://orcid.org/0000-0003-2864-066X
Andrey E. Donnikov, PhD, Head of the Laboratory of Molecular Genetic Methods, V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, 117997, Russia, Moscow, Oparin str., 4, donnikov@dna-technology.ru, https://orcid.org/0000-0003-3504-2406
Corresponding author: Victoria G. Averkova, buch1202@mail.ru



