Value of embryonic mitochondrial DNA in predicting the effectiveness of assisted reproductive technologies
Research evidence suggests that a sufficient level of mitochondrial DNA (mtDNA) in cumulus cells and oocytes is associated with the high potential of female germ cells for fertilization and is an essential factor contributing to the pre-implantation embryonic development. Several studies have shown that a reduced mtDNA copy number is associated with a higher pregnancy rate.Nepsha O.S., Kulakova E.V., Ekimov A.N., Drapkina Yu.S., Makarova N.P., Kraevaya E.E., Kalinina E.A.
Objective: To investigate if mtDNA content in trophectoderm (TE) cells during preimplantation genetic testing for aneuploidy (PGT-A) could be a new marker for an embryo's implantation potential.
Materials and methods: The mtDNA content of trophectoderm cells was analyzed in 244 euploid embryos after OGT-A using the NGS technique.
Results: Analysis of euploid embryo transfer results showed that the mtDNA content in TE blastocyst cells was not associated with ART outcomes. Also, there was no association between the mtDNA level and embryo sex. A relationship was noted between the degree of early blastocyst expansion and mtDNA level. Blastocysts with TEs corresponding to excellent grade A had higher mtDNA content. The mtDNA copy number was significantly lower in embryos biopsied on day six after fertilization compared to biopsied embryos on day 5 of culture. The mtDNA content in all TE cells in euploid embryos was statistically insignificantly correlated with maternal age.
Conclusion: The study findings suggest that mtDNA content in TE cells is not predictive for the embryo implantation potential and ART effectiveness. It should be emphasized that blastocysts with an increased mtDNA levels in TE cells was associated with the birth of healthy children in the ART program. These results confirm that the quantitative mtDNA content is one of the critical factors associated with blastocyst development and depends on the day of embryo culture.
Keywords
Assisted reproductive technology (ART) is considered one of the most effective treatments for infertility today [1]. The development of new modified stimulation protocols and the improvement of the embryological stage has resulted in the increased effectiveness of ART. However, the effectiveness of one attempt does not exceed 40%, and the live birth rate is about 33.3% in patients aged <37 [2]. Many studies have shown that successful implantation requires a functional communication between the receptive and responsive endometrium and a quality embryo with maximum implantation potential. The Gardner blastocyst grading system is used to assess blastocyst stage embryos and their capacity for implantation [3]. However, this grading system remains subjective, and not all embryos of good or excellent morphological quality are successfully implanted [4]. Due to current advancements of one embryo transfer, various technologies have been developed and proposed to improve embryo selection [5]. One of the widely used techniques to optimize the selection of an embryo with a high implantation potential is preimplantation genetic testing for aneuploidy (PGT-A) [6]. Nevertheless, despite the indisputable advantages of PGT-A in terms of increasing the effectiveness of ART programs, the pregnancy rate in ART cycles during euploid blastocyst transfer is about 45–50% [7]. Besides, the onset of clinical pregnancy in the euploid embryo transfer cycle does not guarantee the birth of a healthy child. It does not exclude the presence of balanced chromosome aberrations and single-gene mutations, which may be associated with the development of congenital malformations.
Embryo implantation failure in ART can be associated with the embryo quality and various maternal factors [8].
In recent years, scientific research has achieved a significant breakthrough in the study and understanding of early embryogenesis and embryo implantation at the molecular level and methods for assessing the embryo quality and implantation potential. It has been suggested that mitochondrial DNA (mtDNA) copy numbers in blastocyst biopsy specimens can serve as a biomarker for embryo viability and may be used for embryo selection.
Several studies have shown that euploid embryos with lower mtDNA copy number are more likely to lead to pregnancy [9, 10].
Mitochondria are involved in regulating multiple essential cellular processes, such as adenosine triphosphate (ATP) production, regulation of apoptosis, calcium signaling, reactive oxygen species, the cycle of pyruvate, and citric acid formation, heme and steroid synthesis, and hormonal signal transmission [11]. Mitochondria are maternally inherited [12], since during fertilization, the mitochondria of the spermatozoa are destroyed, and all the mitochondria of the zygote are provided by the fertilized oocyte. Individual organelles can contain more than one copy of the mitochondrial genome, and each cell contains many mitochondria. The mtDNA content of each human cell type is highly variable, with the extremes being the spermatozoa that include only a few copies and mature oocytes with up to several hundred thousand mtDNA copies [13].
Considering that mitochondrial biogenesis begins during embryo implantation, and mtDNA replication is initiated at the blastocyst stage, a sufficient level of mtDNA in cumulus cells and oocytes determines both the increased potential of female germ cells for fertilization and is one of the critical factors contributing to preimplantation embryo development [14, 15]. The results of several studies have shown that a decrease in the mtDNA content in oocytes is associated with impaired fertilization and further embryo development [16]. Cecchino et al. and Hashimoto et al. showed that the mtDNA content remains stable during the initial mitotic divisions since mitochondria and oocyte mtDNA are evenly distributed to the newly formed blastomeres depending on their volume [17, 18]. In a blastocyst-stage embryo, the mtDNA content in TE cells gradually decreases due to the larger cell size and a greater number of cell divisions.
In 2015, two independent groups of authors proposed for the first time to use quantitative assessment of the mtDNA copy number as a biomarker of euploid embryo quality and implantation ability [9, 10]. Scientists have shown that euploid blastocysts that did not lead to pregnancy in the ART had significantly higher mtDNA content in TE cells than successfully implanted embryos. In studies, a mtDNA quantity threshold was obtained, and embryos with an increased level of mtDNA did not lead to pregnancy, in contrast to embryos with a lower level of mtDNA. After the publication of the results, the quantitative mtDNA determination in embryos attracted the attention of researchers and triggered additional studies, which confirmed the initial data [19–21]. Nevertheless, other authors reported conflicting results, which cast doubt on the possibility of using mtDNA as a marker of the quality and implantation potential of the embryo [22–26].
Given the controversial results of previous research, our study aimed to investigate mtDNA content in trophectoderm cells during PGT-A as a new marker for an embryo's implantation potential and analyze outcomes of ART with euploid blastocyst transfer after PGT-A.
Materials and methods
This retrospective study was conducted at the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, V.I. Kulakov NMRC for OG&P of Ministry of Health of Russia. The study analyzed the outcomes of ART of 187 married couples. We analyzed 244 non-mosaic euploid blastocysts obtained as a result of the ART program with PGT-A. All couples provided signed informed consent. The outcomes of 218 selective embryo transfers in cryopreserved cycles were analyzed; 21, 2, 1, and 166 patients underwent 2, 3, 4, and 1 cryopreserved cycles, respectively.
All patients started controlled ovarian stimulation on day 2–5 of their menstrual cycle using the ovarian stimulation protocol with gonadotropin-releasing hormone antagonists and recombinant follicle-stimulating hormone or human menopausal gonadotropin.
Human chorionic gonadotropin (hCG) was used as a trigger. Ovulation was triggered by a single hCG injection of 6,000–10,000 IU intramuscularly when the follicles are 15–18 mm in size.
The transvaginal ovarian puncture was performed 36 hours after the ovulation trigger injection to retrieve oocytes, followed by oocyte quality assessment. Immediately after follicular fluid aspiration, oocyte-cumulus complexes were identified, and oocyte maturity was evaluated under a stereomicroscope on the heated surface of a sterile laminar box. Fertilization of oocytes was carried out by intracytoplasmic sperm injection (ICSI), after which the fertilized cells were transferred to the culture medium (CSCM, Irvine Sc., USA) for further embryo culture. Fertilization is assessed 14–18h after injection by the presence of 2 pronuclei in the oocyte. All stages of embryo culture were carried out in the SOOK MultiGas Incubator (Ireland) in 25 μL drops under oil (Irvine Sc., USA). The medium CSCM, Irvine Sc., USA) was not changed during five days of embryo culture. On day 5 or 6 after fertilization, a TE biopsy was performed, and embryos were cryopreserved. The resulting TE cells were transferred to Eppendorf tubes containing lysis buffer for PGT-A using next-generation sequencing. The PGT-A procedure consisted of several stages. At the first stage, whole-genome amplification and preparation of the library for application to the chip were carried out. Special molecular barcode labels unique for each sample were attached to the DNA fragments to create a library. Then ion semiconductor sequencing was performed, followed by bioinformatic analysis of the results and preparation of a conclusion based on the data obtained according to the standard PGT-A. The normalization of mtDNA copy number was carried out on autosomes according to the manufacturer's software interface.
Frozen-thawed euploid embryo transfer was performed after 1–2 menstrual cycles based on PGT-A results. Patients received cyclic hormonal therapy starting [estradiol valerate (6 mg/day) on day 4–5 of the menstrual cycle and vaginal micronized progesterone (400-600 mg/day) from day 15–16 of the menstrual cycle]. Patients underwent ultrasound monitoring of the changes in endometrial growth on day 9–10 of the menstrual cycle and on day 15–16 of the cycle for gestagens administration. Embryo transfer was carried out on days 20–21 of the menstrual cycle using a Wallace (Germany) or Cook (Australia) soft embryo transfer catheter. The preliminary thawing of embryos and the management of the post-transfer period were carried out according to the protocols accepted in clinical practice.
The biochemical, ectopic and clinical pregnancy rates, miscarriage, implantation, and live birth rate were assessed as primary clinical outcomes of ART. Pregnancy was detected by measuring serum β-hCG on day 14 after the embryo transfer. Pregnancy was defined by a finding of plasma β-hCG concentration > 35 mU/l. If β-hCG was positive, transvaginal ultrasound was performed to diagnose clinical intrauterine pregnancy on day 21 after embryo transfer. The obtained results were presented in the form of a model in which miscarriage, ectopic pregnancy, and clinical pregnancy rates were combined into one group, "positive implantation result," and the absence of pregnancy after euploid embryo transfer, as well as biochemical pregnancy, corresponded to the group "negative implantation result."
Statistical analysis
Statistical analysis was performed using the IBM SPSS Statistics software version 23.0 (USA) and Microsoft Excel. The distribution of continuous variables was tested for normality using the Kolmogorov–Smirnov test and graphical data analysis. Variables not meeting normality assumptions were compared with a nonparametric Mann–Whitney test. Categorical variables were presented as counts (n) and percentages (%). Quantitative variables showing normal distribution were expressed as means (M) and standard deviation (SD); otherwise, the median (Me) with interquartile range (Q1; Q3) were reported. Blastocyst quality, type of aneuploidy, or culture conditions were considered categorical variables. Evaluation of differences between the days of biopsy, sex of the embryo, outcomes of ART programs (implantation rate and clinical pregnancy rate) in terms of mtDNA level was determined using the Mann–Whitney U-test due to the asymmetric distribution of the response. Associations between woman's age and mtDNA content were tested by Pearson correlation. Kruskal–Wallis test was used for comparing blastocyst expansion and the mtDNA copy number between groups. The critical level of significance when interpreting the results of statistical analysis was considered at p<0.05; for better visual perception, all mtDNA values were multiplied by 10,000.
Results
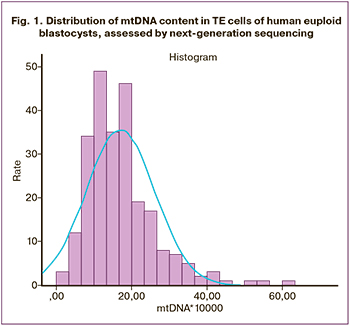
The mtDNA content was determined in all 244 analyzed samples. The distribution of mtDNA copy number in TE cells of euploid embryos showed positive asymmetry (1.508) and prolonged excess (3.568) (Fig. 1). According to the Kolmogorov–Smirnov test result (p<0.001), the distribution of mtDNA did not meet the normality assumption. The Me of the mtDNA copy number and interquartile range were 15.40 (10.67; 20.78). Large scatter of mtDNA copy number was observed with the minimum and the maximum values of 2.06 and 60.70, respectively.
Quantification of mtDNA by maternal age
Maternal age was a continuous variable. The mean age of women at the time of transvaginal puncture was 34.07 (4.08) years (range 26.0–43.0 years). We analyzed the correlation between mtDNA content and maternal age using Pearson's rank correlation. The content of mtDNA in all TE cells of euploid embryos showed a statistically insignificant and very weak correlation with maternal age (Pearson's correlation coefficient r=-0.098, p=0.102).
Quantification of mtDNA and embryonic factors
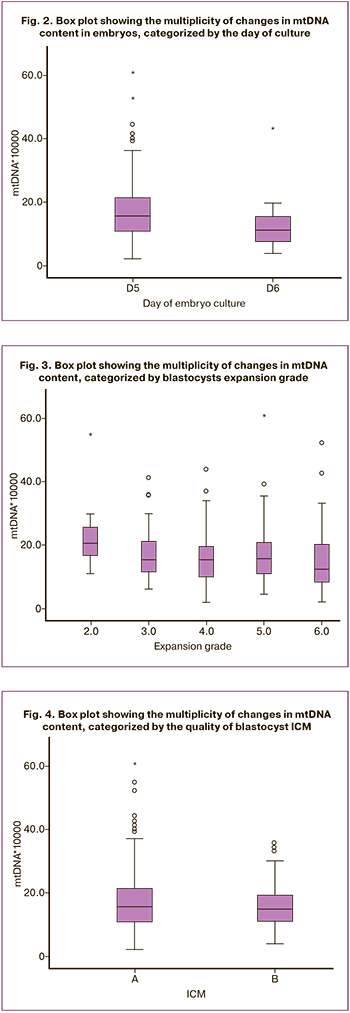
In the present study, we analyzed several embryological factors that could potentially affect embryonic cell mtDNA content. When assessing embryological factors, the relationship between mtDNA content and the day of the biopsy was analyzed. In this study, 215 embryos (88%) were biopsied on day 5 (D5) of culture, 29 (12%) were biopsied on day 6 (D6) after fertilization. D5 embryos had higher mtDNA content than D6 embryos: 15.70 (11.10; 21.80) vs. 11.50 (7.53; 16.55), respectively, p=0.001 (Mann–Whitney test) (Fig. 2).
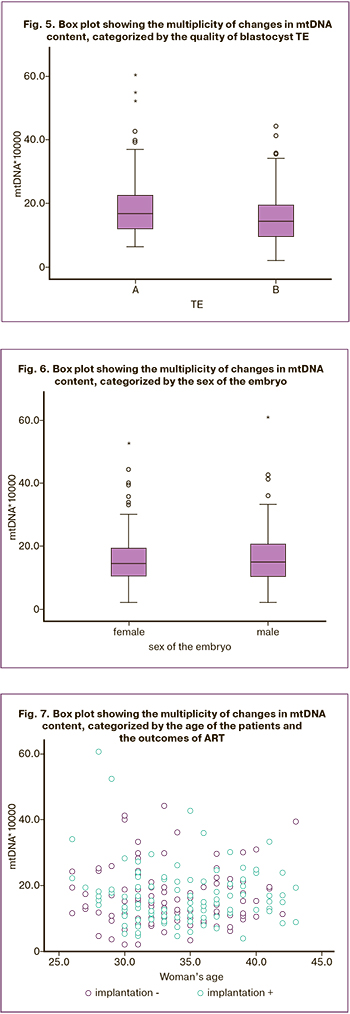
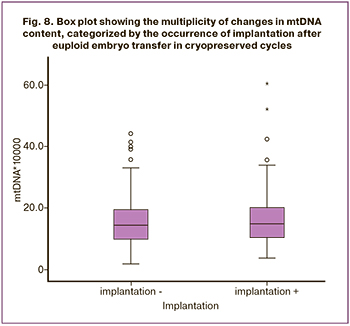
For PGT-A, blastocyst biopsy of good and excellent quality was performed, determined by assessing blastocyst expansion grade (grades 2–6), the quality of the intracellular mass (ICM) (A or B), and TE cells (A or B). A statistically insignificant relationship was found between blastocyst expansion grade and the mtDNA copy number (p=0.178). It is worth noting that there was a relationship between blastocyst expansion grade of early blastocysts (grades 1–2) and an increased mtDNA content, and blastocysts in which the expansion corresponded to grade 6 with a reduced mtDNA content. Pairwise comparison of the two groups using the Mann–Whitney showed a significance level of p=0.041. However, when the Bonferroni correction was applied, the differences between the two groups were not statistically significant (Fig. 3). Moreover, the quality of the blastocyst ICM was not associated with the mtDNA copy number. The number of blastocysts with ICM quality corresponding to grade A was 179 (73.4%), Me 15.60 (10.60; 22.10), grade B – 65 (26.6%), Me 14.70 (10.55; 19.35), p=0.253 (Fig. 4). Comparison of the mtDNA copy number in TE cells showed that blastocysts with TE corresponding to the excellent quality category A (100 blastocysts, 41%) had higher mtDNA values (Me 16.85 (12.12; 22.25) than embryos with TE of good quality B (144 blastocysts, 59%), (Me 14.50 (9.72; 19.60), p=0.007) The Spearman correlation between the quality of TE cells and the mtDNA copy number was -0.174 with significance level p=0.006 (Fig. 5).
We also assessed the effect of the sex of the embryo on mtDNA content. No statistically significant differences were found concerning the sex of the embryo and mtDNA content. The study included 110 XY (45.1%) and 134 XX embryos (54.9%): Me 15.55 (10.60; 20.43) versus 15.30 (10.83, 21.23), respectively, p=0.917 (Fig. 6).
Assessment of mtDNA content depending on ART outcomes
In this study, the mean age of patients was 34.35 (4.31) and 33.47 (3.91) in the group with a positive result and negative result of implantation, respectively, p=0.144 (Fig. 7). The group with a positive result of implantation consisted of 102 patients (46.8%), of which one woman was diagnosed with an ectopic pregnancy, nine patients had a miscarriage at 4–8 weeks of pregnancy, and 92 women had a confirmed clinical pregnancy. In the group with a negative result of implantation, there were 116 patients (53.2%), of whom biochemical pregnancy occurred in 5 women, and 111 patients had a negative β-hCG test result.
There was no significant difference in mtDNA content between the group with a positive and negative result of implantation: Me 15.15 (10.60; 20.63) versus 14.47 (10.23; 19.68), respectively, p=0, 56 (Fig. 8). Outliers corresponding to high levels of mtDNA were recorded in both groups. Information on pregnancy outcomes and live births was collected from 72 out of 92 women with confirmed clinical pregnancy. At the time of manuscript drafting, 16 women had developed pregnancy according to the gestational age, and the live birth rate was 35.6%. In 72 patients who gave birth to children, three embryos had an mtDNA content above 40 units. There was only one embryo with mtDNA content exceeding 40 copies per cell among patients with implantation failure.
Discussion
Despite the availability of many markers of implantation potential and embryo quality, it is not always possible to explain the cause of implantation failure of a morphologically good embryo. In this study, the analysis of mtDNA content in embryos of excellent and good quality was carried out depending on the result of implantation, the qualitative characteristics of the embryo, maternal age, the sex of the embryo, and the day of culture. Analysis of 218 ART program outcomes showed that mtDNA content in human euploid blastocysts did not depend on reproductive outcomes. The results of previous studies on the use of mtDNA as a marker of implantation have also shown conflicting results [9, 10, 23, 24].
The results of two studies published in 2015 [9, 10] demonstrated that lower mtDNA content in TE cells in euploid embryos is a marker of potentially successful blastocyst implantation in ART. Diez-Juan et al. determined the mtDNA quantity threshold in embryos, the excess of which never led to the onset of pregnancy after such an embryo transfer [10]. Another group of authors has developed a scoring scale for assessing an embryo capacity for implantation depending on the quantitative level of mtDNA. In 2017, teams of researchers in collaboration with Fragouli et al. published two more studies [19, 20], reporting the mtDNA quantity threshold predicting IVF failure after euploid embryo transfer.
The early development of the embryo depends on a series of coordinated cell divisions regulated by ATP. During the blastocyst stage, ATP production is increased to provide the cells with the energy required for further differentiation and development of the embryo and maintain the processes needed for implantation [9]. The increase in mtDNA content in non-viable embryos is most likely due to the need for an additional source of energy under conditions of cellular stress. The hypothesis of an increase in mtDNA level in embryos under stress is evidenced by the fact that mtDNA content in aneuploid embryos is much higher than in euploid ones [26, 27]. There is a theory of a silent embryo. A viable embryo has a lower and quieter metabolism, in contrast to embryos under cellular stress conditions and a reduced development potential [9, 10, 26]. As an alternative hypothesis, the authors suggested that some embryos increase mtDNA content to compensate for mitochondrial deficiency associated with mutations in the mitochondrial genome [20]. Liedo et al. described the relationship between mitochondrial genome mutations and an increased mtDNA content, which led to a decrease in implantation rates of embryos with heteroplasmic variants of mtDNA [21].
This study's findings are not entirely consistent with the conclusions mentioned above. Our data suggest that mtDNA level was not associated with implantation rates in the ART program. Besides, this study did not find a mtDNA quantity threshold predicting implantation failure. Thus, the results of this study are consistent with those of Victor et al. that the transfer of euploid embryos with a high mtDNA copy number can lead to the birth of healthy children [24, 25]. According to a study by Treff et al. [23], when transferring two euploid embryos of different sexes to 69 patients and successful implantation of one of the embryos, no difference was found in the relative mtDNA copy number in embryos that led to pregnancy and in the group of embryos with implantation failure. We analyzed the mtDNA level in embryos obtained from the same married couple to determine whether mtDNA content is a predictor of embryo implantation potential. The study included patients with pregnancy failure in one cycle after embryo transfer, and in the subsequent cycle, the transfer of a thawed embryo led to pregnancy (39 women, 78 transferred embryos). There were no changes in the relative mtDNA copy number in embryos obtained from the same parents and having different implantation potential.
The study findings showed no differences in mtDNA content in embryos obtained in the groups of women of older reproductive age and younger patients [9, 25]. Statistically significant changes were found only between patients aged 21–22 and 42–48 years, which can be explained by pronounced dysfunctional changes in embryos obtained from patients of older reproductive age [19].
This study showed that mtDNA copy number was significantly lower in embryos biopsied on D6 compared to biopsied embryos on D5, which is consistent with the results of other authors [21, 23]. Thus, mitochondria's content in the preimplantation embryo cell depends on the number of cell divisions preceding the biopsy and may reflect a decrease in the relative content of mtDNA per cell as a result of cell division in the growing blastocyst. It is important to note that upon reaching the blastocyst stage, mtDNA replication factors (POLGA, POLGB, and TFAM) are activated, which leads to an increase in the total mtDNA copy number [14]. According to Wu F.S.-Y. et al. [28], the main reason for culturing embryos up to D6 is to give lagging embryos more time to develop. Therefore, the total cell mass of blastocysts D5 and D6 should be approximately similar. The authors suggested that the oocytes from which the D6 blastocysts were obtained initially had a suboptimal mtDNA content, which led to an energy deficit and required additional time to complete the blastulation process. For the formation of the blastocyst cavity, high levels of ATP are needed, and the morula, which has a reduced mitochondrial function, is unable to form a blastocoel, which leads to a delay in its development [10, 28]. Thus, D6 blastocysts have a slowed down metabolic activity and growth due to internal or external stress, which forces cells to increase mtDNA content to compensate for the energy deficit.
This study found that embryos with excellent morphological quality of TE cells (A) and expanding blastocysts contained more mtDNA copies per cell than embryos with good quality TE (B). Several studies have shown that embryos with excellent morphological characteristics contain less mtDNA [10, 22, 23]. Our findings confirm that expansion grade and mitogenic activity of the blastocyst affect mtDNA content.
The discrepancy between the results of similar studies among different authors is possibly related to the methodological differences in determining the mtDNA content in embryos. It cannot be ruled out that measurements using next-generation sequencing of genome-wide amplification products obtained using polymerase chain reaction may not be accurate enough to provide comprehensive results. The data obtained by measuring the content of mtDNA using real-time polymerase chain reaction with specific fluorescent probes previously showed that the mtDNA copy number in TE of aneuploid blastocysts is statistically higher than that of euploid blastocysts [29].
Conclusion
Our study findings suggest that mtDNA content in TE cells does not predict embryo implantation potential and the effectiveness of the ART program. It should be emphasized that blastocysts with significant amounts of mtDNA in TE cells were associated with the birth of healthy children in the ART program. Moreover, it is not recommended to use an arbitrary mtDNA quantity threshold since different clinics obtained different cutoffs for the quantitative mtDNA content in cells, above which all embryos failed to implant after a euploid embryo transfer in the ART program. This study confirms that blastocyst cells on day five after fertilization contain a significantly higher amount of mtDNA than that on day 6 of culture, and mtDNA quantity is one of the critical factors associated with blastocyst development rate.
To better understand the role of mtDNA in early embryogenesis and the clinical value of this marker, further studies are needed, investigating the relationship between euploid embryo morphology and mtDNA quantity both in the general group of patients and in embryos obtained from the same married couples. Besides, the optimization of existing or the development of new, more accurate methods for studying mtDNA content in embryos is required.
References
- World Health Organization. Sexual and reproductive health. 2020. Available at: https://www.who.int/reproductivehealth/topics/infertility/perspective/en/
- SART: Society for Reproductive Technology. National Summary Report. Final National Summary Report for 2016. Available at: https://wwwsartcorsonlinecom/rptCSR_PublicMultYearaspx?reportingYear=2016
- Gardner D.K., Schoolcraft W.B. In vitro culture of human blastocysts. In: Jansen R., Mortimer D., eds. Towards reproductive certainty: fertility and genetics beyond. Parthenon Press, Carnforth; 1999: 378-88.
- Bromer J.G., Ata B., Seli M., Lockwood C.J., Seli E. Preterm deliveries that result from multiple pregnancies associated with assisted reproductive technologies in the USA: a cost analysis. Curr. Opin. Obstet. Gynecol. 2011; 23(3): 168-73. https://dx.doi.org/10.1097/GCO.0b013e32834551cd.
- Timofeeva A.V., Chagovets V.V., Drapkina Yu.S., Makarova N.P., Kalinina E.A., Sukhikh G.T. Cell-free, embryo-specific sncRNA as a molecular biological bridge between patient fertility and IVF efficiency. Int. J. Mol. Sci. 2019; 20(12): 2912. https://dx.doi.org/10.3390/ijms20122912.
- Кулакова Е.В., Калинина Е.А., Трофимов Д.Ю., Макарова Н.П., Хечумян Л.Р., Дударова А.Х. Вспомогательные репродуктивные технологии у супружеских пар с высоким риском генетических нарушений. Преимплантационный генетический скрининг. Акушерство и гинекология. 2017; 8: 21-7. [Kulakova E.V., Kalinina E.A., Trofimov D.Yu., Makarova N.P., Khechumyan L.R., Dudarova A.Kh. Assisted reproductive technologies in couples with a high risk of genetic disorders. Preimplantation genetic screening. Obstetrics and gynecology. 2017; 8: 21-7. (in Russian)]. https://dx.doi.org/10.18565/aig.2017.8.21-7.
- Долгушина Н.В., Коротченко О.Е., Бейк Е.П., Абдурахманова Н.Ф., Ильина Е.О., Кулакова Е.В. Клинико-экономический анализ эффективности преимплантационного генетического скрининга у пациенток позднего репродуктивного возраста. Акушерство и гинекология. 2017; 11: 56-61. [Dolgushina N.V., Korotchenko O.E., Beyk E.P., Abdurakhmanova N.F., Ilyina E.O., Kulakova E.V. Clinical and economic analysis of the effectiveness of preimplantation genetic screening in patients of advanced maternal age. Obstetrics and gynecology. 2017; 11: 56-61. (in Russian)]. https: //dx.doi.org/10.18565/aig.2017.11.56-61.
- Tarín J.J., García-Pérez M.A., Hamatani T., Cano A. Infertility etiologies are genetically and clinically linked with other diseases in single meta-diseases. Reprod. Biol. Endocrinol. 2015; 13: 31. https://dx.doi.org/10.1186/s12958-015-0029-9.
- Fragouli E., Spath K., Alfarawati S., Kaper F., Craig A., Michel C.-E. et al. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet. 2015; 11(6): e1005241. https://dx.doi.org/10.1371/journal.pgen.1005241.
- Diez-Juan A., Rubio C., Marin C., Martinez S., Al-Asmar N., Riboldi M. et al. Mitochondrial DNA content as a viability score in human euploid embryos: less is better. Fertil. Steril. 2015; 104(3): 534-41.e1. https://dx.doi.org/0.1016/j.fertnstert.2015.05.022.
- Harvey A.J. Mitochondria in early development: linking the microenvironment, metabolism and the epigenome. Reproduction. 2019; 157(5): R159-R179. https://dx.doi.org/10.1530/REP-18-0431.
- Vaught R.C., Dowling D.K. Maternal inheritance of mitochondria: implications for male fertility? Reproduction. 2018; 155(4): R159-R168. https://dx.doi.org/10.1530/REP-17-0600.
- Archer S.L. Mitochondrial dynamics – mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 2013; 369(23): 2236-51. https://dx.doi.org/10.1056/NEJMra1215233.
- Ciesielski G.L., Oliveira M.T., Kaguni L.S. Animal mitochondrial DNA replication. Enzymes. 2016; 39: 255-92. https://dx.doi.org/10.1016/bs.enz.2016.03.006.
- Popov L.-D. Mitochondrial biogenesis: an update. J. Cell. Mol. Med. 2020; 24(9): 4892-9. https://dx.doi.org/10.1111/jcmm.15194.
- Королькова А.И., Мишиева Н.Г., Мартазанова Б.А., Бурменская О.В., Веюкова М.А., Екимов А.Н., Трофимов Д.Ю., Абубакиров А.Н. Значимость копийности митохондриальной ДНК в клетках кумулуса пациенток позднего репродуктивного возраста. Акушерство и гинекология. 2019;10: 108-14. [Korolkova A.I., Mishieva N.G., Martazanova B.A., Burmenskaya O.V., Veyukova M.A., Ekimov A.N., Trofimov D.Yu., Abubakirov A.N. The significance of mitochondrial DNA copy number in cumulus cells in patients of advanced reproductive age. Obstetrics and gynecology. 2019; 10: 108-14. (in Russian)]. https://dx.doi.org/10.18565/aig.2019.10.108-114.
- Cecchino G.N., Garcia-Velasco J.A. Mitochondrial DNA copy number as a predictor of embryo viability. Fertil. Steril. 2018: 111(2): 205-11. https://dx.doi.org/10.1016/j.fertnstert.2018.11.021.
- Hashimoto S., Morimoto N., Yamanaka M., Matsumoto H., Yamochi T., Goto H. et al. Quantitative and qualitative changes of mitochondria in human preimplantation embryos. J. Assist. Reprod. Genet. 2017; 34(5): 573-80. https://dx.doi.org/10.1007/s10815-017-0886-6.
- Ravichandran K., McCaffrey C., Grifo J., Morales A., Perloe M., Munné S. et al. Mitochondrial DNA quantification as a tool for embryo viability assessment: retrospective analysis of data from single euploid blastocyst transfers. Hum. Reprod. 2017; 32(6): 1282-92. https://dx.doi.org/10.1093/humrep/dex070.
- Fragouli E., McCaffrey C., Ravichandran K., Spath K., Grifo J.A., Munné S. et al. Clinical implications of mitochondrial DNA quantification on pregnancy outcomes: a blinded prospective non-selection study. Hum. Reprod. 2017; 32(11): 2340-7. https://dx.doi.org/10.1093/humrep/dex292.
- Lledo B., Ortiz J.A., Morales R., Garcia-Hernandez E., Ten J., Bernabeu A. et al. Comprehensive mitochondrial DNA analysis and IVF outcome. Hum. Reprod. Open. 2018; 2018(4): hoy023. https://dx.doi.org/10.1093/hropen/hoy023.
- Klimczak A.M., Pacheco L.E., Lewis K.E., Massahi N., Richards J.P., Kearns W.G. et al. Embryonal mitochondrial DNA: relationship to embryo quality and transfer outcomes. J. Assist. Reprod. Genet. 2018; 35(5): 871-7. https://dx.doi.org/10.1007/s10815-018-1147-z.
- Treff N.R., Zhan Y., Tao X., Olcha M., Han M., Rajchel J. et al. Levels of trophectoderm mitochondrial DNA do not predict the reproductive potential of sibling embryos. Hum. Reprod. 2017; 32(4): 954-62. https://dx.doi.org/10.1093/humrep/dex034.
- Victor A., Griffin D., Dardner K.G., Brake A., Zouves C., Barnes F. et al. Births from embryos with highly elevated levels of mitochondrial DNA. Reprod. Biomed. Online. 2019; 39(3): 403-12. https://dx.doi.org/10.1016/j.rbmo.2019.03.214.
- Victor A., Brake A.J., Tyndall J.C., Griffin D.K., Zouves C.G., Barnes F.L. et al. Accurate quantitation of mitochondrial DNA reveals uniform levels in human blastocysts irrespective of ploidy, age, or implantation potential. Fertil. Steril. 2017; 107(1): 34-42.e3. https://dx.doi.org/10.1016/j.fertnstert.2016.09.028.
- Lee Y.-X., Chen C.-H., Lin S.-Y., Lin Y.-H., Tzeng C.-R. Adjusted mitochondrial DNA quantification in human embryos may not be applicable as a biomarker of implantation potential. J. Assist. Reprod. Genet. 2019; 36(9): 1855-65. https://dx.doi.org/10.1007/s10815-019-01542-6.
- Shang W., Zhang Y., Shu M., Wang W., Ren L., Chen F. et al. Comprehensive chromosomal and mitochondrial copy number profiling in human IVF embryos. Reprod. Biomed. Online. 2018; 36(1); 67-74. https://dx.doi.org/10.1016/j.rbmo.2017.10.110.
- Wu F.S.Y., Weng S.P., Shen M.S., Ma P.C., Wu P.K., Lee N.C. Suboptimal trophectoderm mitochondrial DNA level is associated with delayed blastocyst development. J. Assist. Reprod. Genet. 2021; 38(3): 587-94. https://dx.doi.org/10.1007/s10815-020-02045-5.
- Королькова А.И., Мишиева Н.Г., Мартазанова Б.А., Бурменская О.В., Екимов А.Н., Трофимов Д.Ю., Веюкова М.А., Кириллова А.О., Абубакиров А.Н. Повышение эффективности программ ЭКО на основании определения копийности митохондриальной ДНК в трофэктодерме эмбрионов. Акушерство и гинекология. 2019; 3: 98-104. [Korolkova A.I., Mishieva N.G., Martazanova B.A., Burmenskaya O.V., Ekimov A.N., Trofimov D.Yu., Veyukova M.A, Kirillova A.O., Abubakirov A.N. Increasing the effectiveness of IVF programs by determining mitochondrial DNA copy number in embryonic trophectoderm. Obstetrics and gynecology. 2019; 3: 98-104. (in Russian)]. https://dx.doi.org/10.18565/aig.2019.3.98-104.
Received 03.06.2021
Accepted 23.06.2021
About the Authors
Oksana S. Nepsha, Ph.D. (Biol. Sci.), Researcher at the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, V.I. Kulakov NMRC for OG&P,Ministry of Health of Russia, nepsha@oparina4.ru, https://orcid.org/0000-0002-9988-2810, 117997, Russia, Moscow, Academician Oparin str., 4.
Elena V. Kulakova, Ph.D., Senior Researcher at the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, V.I. Kulakov NMRC for OG&P,
Ministry of Health of Russia, e_kulakova@oparina4.ru, https://orcid.org/0000-0002-4433-4163, 117997, Russia, Moscow, Academician Oparin str., 4.
Alexey N. Ekimov, Clinical Geneticist at the Molecular Genetics Laboratory, V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, a_ekimov@oparina4.ru,
https://orcid.org/0000-0001-5029-0462, 117997, Russia, Moscow, Academician Oparin str., 4.
Yulia S. Drapkina, Ph.D., Obstetrician-Gynecologist at the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, V.I. Kulakov NMRC for OG&P,
Ministry of Health of Russia, yu_drapkina@oparina4.ru, https://orcid.org/0000-0002-0545-1607, 117997, Russia, Moscow, Academician Oparin str., 4.
Natalia P. Makarova, Dr. Biol. Sci., Senior Researcher at the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, np_makarova@oparina4.ru, https://orcid.org/0000-0003-8922-2878, 117997, Russia, Moscow, Academician Oparin str., 4.
Elizaveta E. Kraevaya, Ph.D., Junior Researcher at the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, V.I. Kulakov NMRC for OG&P,
Ministry of Health of Russia, e_kraevaya@oparina4.ru, 117997, Russia, Moscow, Academician Oparin str., 4.
Elena A. Kalinina, Dr. Med. Sci., Professor, Head of the B.V. Leonov Department of Assisted Technologies for the Treatment of Infertility, V.I. Kulakov NMRC for OG&P,
Ministry of Health of Russia, e_kalinina@oparina4.ru, https://orcid.org/0000-0002-8922-2878, 117997, Russia, Moscow, Academician Oparin str., 4.
Authors' contributions: Kulakova E.V., Makarova N.P. – manuscript editing; Nepsha O.S., Drapkina Yu.S., Kraevaya E.E. – collection and review of the relevant literature; Ekimov A.N. – preimplantation genetic testing by next-generation sequencing; Makarova N.P. – embryo cultivation, embryo trophectoderm biopsy, embryo cryopreservation, editing and final approval of the manuscript; Kalinina E.A. – editing and final approval of the manuscript.
Conflicts of interest: The authors have no conflicts of interest to declare.
Funding: There was no funding for this study.
Patient Consent for Publication: All patients provided informed consent for the publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator.
For citation: Nepsha O.S., Kulakova E.V., Ekimov A.N., Drapkina Yu.S., Makarova N.P., Kraevaya E.E., Kalinina E.A. Value of embryonic mitochondrial DNA in predicting the effectiveness of assisted reproductive technologies.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2021; 11: 125-134 (in Russian)
https://dx.doi.org/10.18565/aig.2021.11.125-134



