Whole exome sequencing in couples with unexplained infertility (pilot study)
Aim: To conduct a pilot study of whole exome sequencing in couples with unexplained infertility.Kirakosyan E.V., Pomerantseva E.A., Pavlovich S.V.
Materials and methods: Whole exome sequencing was performed using DNA from peripheral blood samples obtained from six married couples (women and men). The samples were purposefully selected in the group of unexplained infertility on the basis of clinical and anamnestic findings, laboratory and instrumental parameters, and the results of using assisted reproductive technologies. Assessment of clinical significance (pathogenicity) of the identified variants was based on the American College of Medical Genetics and Genomics (ACMG) guidance for the interpretation of sequence variants and the recommendations of the Russian Society of Medical Geneticists for interpretation of the obtained data using next-generation sequencing.
Results: Whole exome sequencing in 6 married couples with unexplained infertility showed no pathogenic/likely pathogenic variants related to patients phenotype, as well as incidental (secondary) findings according to ACMG list of genes.
Conclusion: Whole exome sequencing in the most typical patients with unexplained infertility found no variants, which are known to be associated with infertility.
Keywords
Unexplained infertility (UI) is a clear clinical model of reproductive system disorder, where despite the normal structure and function of the reproductive system, pregnancy does not occur [1–3].
According to the published data, there are few studies in the world, which are devoted to UI, and their level of evidence is considered low. International consensus development study established top ten priorities for future infertility research, including UI [4].
Previously we have obtained the data showing that in cases of UI, the number of oocytes was sufficient for in vitro fertilization (IVF) programs, the frequency of fertilization was normal. However, the blastulation rate was low mainly due to the fact that embryo stopped developing within 3 days of culture, that was a sign of impaired early embryogenesis and, therefore, an indication to use IVF for getting pregnant [5].
The range of the studies on the human reproductive system is broadening, and currently includes high-tech methods aimed to obtain genetic, epigenetic and other information about reproductive system disorders [6].
Whole exome sequencing is one of next generation sequencing (NGS) methods, that helps immediately to decipher exon structures, that are DNA sequences encoding human proteins [7].
Whole exome sequencing identifies the variants of known genes, as well as discovers new genetic features and patterns that may be associated with the disease under study. Also, whole exome sequencing can serve as a research tool in the absence of a hypothesis, that is, a preliminary idea of genes, signaling cascades, or metabolic pathways, that altered in the body of patient with UI. In this case, the study does not include familial, but sporadic cases of the disease with a similar clinical picture [8, 9].
Performance of whole exome sequencing in married couples with UI will make it possible to check the presence of variants, the role of which is being discussed in scientific researches and detect new features and patterns of genotype in patients with UI [10]. Thus, poor oocyte quality in infertility can be due to gene variants PATL2, TUBB8, WEE2 and PAD16: alteration of gene expression [OMIM gene ID: 614661, OMIM phenotype: OOMD4 (oocyte maturation defect), OMIM phenotype ID: 617743, autosomal recessive inheritance] PATL2 [15q21.1] and [OMIM gene ID: 616768.0001, OMIM phenotype: OOMD2, OMIM phenotype ID: 616780, autosomal dominant inheritance] TUBB8 [10p15.3] and leads to oocyte maturation arrest at metaphase phase II (MII); [OMIM gene ID: 614084.0001-614084.0003, OMIM phenotype: OOMD5, OMIM phenotype ID: 617996, autosomal recessive inheritance] WEE2 [7q34] – low fertilization ability of oocytes and embryo development; [OMIM gene ID: 610363.0001-610363.0005, OMIM phenotype: PREMBL2 (preimplantation embryonic lethality), OMIM phenotype ID: 617234, autosomal recessive inheritance] PAD16 [1p36.13] and early embryonic arrest. ZP1, ZP3, ZP2, TLE6 genes can also be associated with oocyte maturation defects and preimplantation embryonic lethality [11, 12].
During performance of whole exome sequencing other medically significant variants may be detected as random findings in addition to the main information. Due to this, ethical issues arise about the need and amount of information to be provided to the patient. The specialists of the American College of Medical Genetics and Genomics (ACMG) have published the list of genes and recommended to inform the patients about finding their variants for the purpose of early diagnosis of the disease. However, currently there is no a single solution to the ethical problem of whole exome sequencing [13, 14].
At present, indication for genetic testing in patients with UI is not regulated by the clinical guidelines and is performed empirically in cases of abnormally small number of mature oocytes, low fertilization rate, early embryonic arrest in IVF programs.
The objective of the study was to perform whole exome sequencing in couples with unexplained infertility.
Materials and methods
The diagnosis of unexplained infertility in couples is made when standard infertility testing does not find a cause of infertility in patients with regular, ovulatory menstrual cycle, tubal patency, no pathological endometrial changes, normal sperm count, and no pathology was found during laparoscopy and hysteroscopy.
A pilot study of whole exome sequencing using DNA from peripheral blood samples obtained from six married couples (women and men) was performed in the Department of Clinical Genetics of the Institute of Reproductive Genetics, the National Medical Research Center for Obstetrics, Gynecology and Perinatology named after Academician V.I. Kulakov of the Ministry of Health of Russia (further referred to as the “Center”). The women and men were and methodically chosen in the group of prospective study of patients with unexplained infertility (n=45) to rule out suspected genetic causes of infertility in couples with UI [3].
The patients were chosen for whole exome sequencing according to the following criteria:
- the diagnosis of infertility was made before the patients reached 35 years of age;
- duration of infertility ≥ 5 years;
- uncomplicated somatic and gynecological anamnesis;
- normal level of hormones in blood;
- male partner with normozoospermiaа;
- normal female (46,XX) and male (46ХУ) karyotypes;
- absence of ultrasound features of pathology in the organs and systems;
- failed attempts of intrauterine insemination;
- 3 or more failed attempts of IVF with registration of impairment of embryonic development or absence of embryos.
Assessment of clinical significance (pathogenicity) of the identified variants was based on the American College of Medical Genetics and Genomics (ACMG) guidance for the interpretation of sequence variants and the recommendations of the Russian Society of Medical Geneticists for interpretation of the obtained data using next-generation sequencing [15, 16].
In 6 couples with UI, who were included in the study, 25 ml of peripheral blood was obtained from cubital vein. All patients have signed voluntary informed consent to participate in the study. DNA sequencing was performed using NovaSeq 6000 platform for whole genome sequencing (WES). The Illumina TruSeq Exome Enrichment kit was used for enrichment of target DNA fragments.
Sequencing data analysis was carried out using an algorithm that included aligning reads to reference genome sequence hg38, variant calling and quality-based filtering. All filtered variants were annotated with the Ensembl Variant Effect Predictor (VEP) using a number of algorithms (SIFT, PolyPhen-2, SpliceAI) for variant effect prediction [17].
The genome aggregation database (gnomAD) was used to assess frequency of detected variants in population [18]. Data bases OMIM, ClinVar, LOVD, RUSeq and other specialized data bases were used to assess clinical relevance of the detected variants [19–23].
We considered the variants that could be associated with infertility in couples and incidental findings according to ACMG list of genes for reporting about secondary findings with the exception of undescribed variants in the TTN gene leading to loss of protein function [13, 14, 24].
It should be noted that the significance of the detected variants eventually can be changed with evidence and clinical data accumulation.
Limitations: the method does not allow detection of insertions and deletions of lengths more than 10 pairs of nucleotides, variants in intronic regions (with the exception of canonical splice site variants), repeat length variations (including triplet repeat length). The method is not intended to determine cis/trans status for pairs of heterozygote sequence variants, as well as assess the level of methylation, detect chromosomal rearrangements, polyploidy, mosaic variants [15].
Results
Whole exome sequencing (WES) in 6 married couples with unexplained infertility showed no pathogenic variants related to patient phenotype; no likely pathogenic variants related to patient phenotype; no incidental (secondary) findings according to ACMG list of genes. In two married couple, variants of unclear clinical significance were detected, as described below.
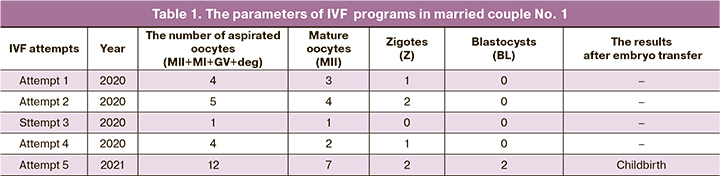
Married couple No. 1: B., aged 27 years (female), S., aged 36 years (male), Moscow, nationality: the Russians. During 6 years of sexual activity without contraception pregnancy did not occur. From 2017 to 2021, the patients repeatedly underwent diagnostic examinations. The cause of inferility was not identified. In 2018–2020, the couple underwent fertility treatment: intrauterine insemination (IUI) two times, two IVF programs: natural cycle IVF and stimulated cycle with gonadotropin-releasing hormone agonist (GnRH-a) long protocol. Pregnancy did not occur. In 2020, 3 IVF attempts using semen from the partner and one attempt with donor insemination resulted in embryonic arrest on days 3–4 (Table 1). The couple underwent diagnostic examination in the Center. The diagnosis was made on 03.03.2021: “Unexplained infertility. Recurrent failures after using assisted reproductive technology programs. Early embryonic arrest”. The patients were recommended to undergo IVF program: 12 oocytes were obtained, out of them 7 were mature oocytes, 2 fertilized oocytes – 2 zygotes, 2 blastocysts were formed. Pregnancy occurred and successfully ended with childbirth.
A variant of unclear clinical significance was found in the woman in this married couple: previously undescribed heterozygous variant of nucleotide sequence in follicle-stimulating hormone receptor (FSHR) (2-48963163-A-G), leading to amino acid substitution (p. Tyr553Cys, NM_000145.4). Databases of Functional Predictions dbNSFP, PolyPhen, SIFT classify the variant as pathogenic. Missense variant of the FSHR gene was described, including in patients with autosomal recessive ovarian dysgenesis (OMIM: 233300, ovarian dysgenesis-1) and autosomal dominant ovarian hyperstimulation syndrome (OMIM: 608115, ovarian hyperstimulation syndrome). The variant was not registered in the control samples in the Genome Aggregation Database (gnomAD) and in the genomic data set RUSeq.
Married couple No. 2: K., aged 34 years (f), K., aged 33 years (m), Moscow, nationality: the Russians. During 10 years of sexual activity without contraception pregnancy did not occur. In 2015, the couple was diagnosed with unexplained infertility. Ovarian stimulation and intrauterine insemination was performed. Pregnancy did not occur. From 2017 to 2021, in 3 IVF attempts, a sufficient number of oocytes was obtained (12, 14, 21 oocytes, respectively), but low blastocyst yield (1, 2, 2 blastocysts, respectively), transfer of one blastocyst was in each program and cryotransfer of the remaining blastocysts suitable for transfer according to preimplantation genetic testing for aneuploidy (PGT-A). Pregnancy did not occur (Table 2).
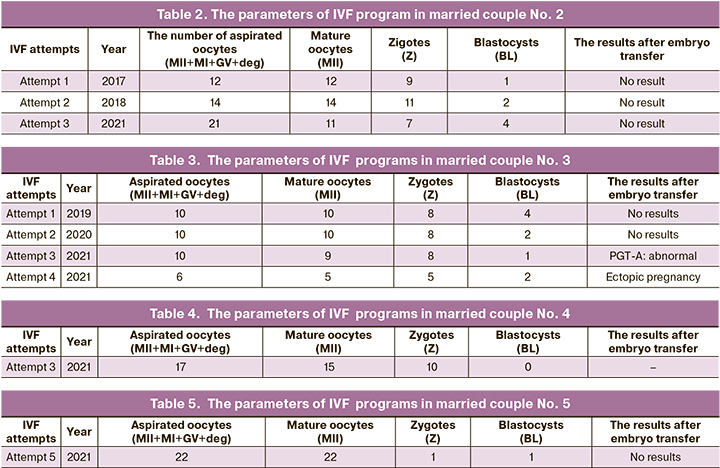
Married couple No. 3: P., aged 39 years (f), P., aged 40 years (m), Kaluzhskaya region, nationality: the Russians. During 5 years of sexual activity without contraception pregnancy did not occur. From 2012 to 2019, the couple repeatedly underwent diagnostic examination. The cause of infertility was not identified. Intrauterine insemination was performed twice: IUI with ovarian stimulation and natural cycle IUI. Pregnancy did not occur. In 2019, the couple was diagnosed with unexplained infertility, and was recommended to undergo IVF program. From 2019 to 2021, there were four IVF attempts. In three of them, there was low blastocyst yield. In the result of last IVF attempt, left ectopic pregnancy was diagnosed at 5–6 weeks. Laparoscopic left tubal ligation was performed (Table 3).
Married couple No.4: B., aged 38 years (f), D., aged 40 years (m), The Republic of North Ossetia – Alania, nationality: the Ossetians. During 5 years of sexual activity without contraception one pregnancy occurred in 2017. Pregnancy loss was at 4 weeks without complications. In 2020, there were 2 IVF attempts, pregnancy did not occur. The third IVF attempt was in 2021; there was a sufficient number of oocytes (out of 17 obtained oocytes 15 were mature oocytes) and zygotes (10), blastocysts were not formed (Table 4).
Married couple No.5: L., aged 34 years (f), L., aged 36 years (m), the Moscow region, nationality: the Russians. During 13 years of sexual activity without contraception pregnancy did not occur. Since 2012 the couple repeatedly underwent diagnostic examination. The cause of infertility was not identified. Ovarian stimulation and intrauterine insemination was performed. Pregnancy did not occur. From 2014 to 2021, there were 4 IVF attemprs in different clinics. The sufficient number of oocytes was obtained (17, 24, 20, 20 oocytes, respectively), and blastocyst formation was adequate (7, 6, 14, 4 blastocysts, respectively), 2 blastocyst stage embryos were transferred in three IVF programs (in the fourth IVF program blastocyst transfer was not performed due to poor embryo quality). PGT-A was not performed due to lack of adequate embryos to be biopsied. In 2021, the couple visited the Center and was recommended to undergo IVF program: 22 oocytes were obtained, out of them 22 were mature oocytes, 1 fertilized oocyte – 1 zygote, 1 blastocyst was formed. Pregnancy did not occur (Table 5). Retrospectively, the diagnosis “Unexplained inferitility. Repeated IVF failures. Oocyte factors?” was made.
A variant of unclear clinical significance was identified in the man – previously described heterozygous nucleotide sequence variant in the GNRHR gene (4-67740682-C-T), leading to amino acid substitution at position 262 of the protein (p.Arg262Gln, NM_000406, rs104893837). The variant was described in compound heterozygous form with other variants and in the homozygous form in patients with hypogonadotropic hypogonadism (OMIM: 146110, Hypogonadotropic hypogonadism 7 without anosmia; OMIM: 138850#0002). The variant was registered in the control sample in gnomAD – the number of mutant alleles was 505 on 282244 chromosomes, including 1 homozygote, and in RUSeq – the number of observations was 4/3376, the frequency of the variant was 0.001185. Based on the totality of information, given the non-detection of the second pathogenic variant in the gene, it could be regarded as a variant with uncertain clinical significance. Based on totality of evidence and given the fact that the second pathogenic variant was not detected in the gene, it could be regarded as a variant with uncertain clinical significance.
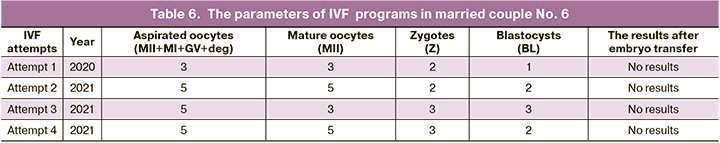
Married couple No.6: A., aged 35 years (f), A., aged 37 years, Moscow, nationality: the Russians. During 6 years of sexual activity without contraception pregnancy did not occur. Since 2019 the couple repeatedly underwent diagnostic examination. The cause of infertility was not identified. From 2020 to 2021, in 3 IVF attempts, the number of oocytes was low (3, 5, 5 oocytes, respectively), and the number of blastocysts was comparable (2, 2, 3 blastocysts, respectfully), single blastocyst transfer, single blastocyst cryotransfer and two times good-quality double blastocyst transfer were performed, respectively. Pregnancy did not occur. PGT-A was not performed due to lack of adequate embryos to be biopsied. In 2021 the diagnosis of unexplained infertility was made. The couple was recommended to undergo the fourth IVF program: 5 oocytes were obtained, out of them 5 were mature oocytes, 3 were fertilized oocytes – 3 zigotes, 2 blastocysts of high quality were formed and transferred. Pregnancy did not occur (Table 6).
Thus, whole exome sequencing in patients with UI did not find known variants that could be associated with infertility. Further research is needed to investigate the molecular mechanisms of impairment in early stages of embryogenesis in patients with UI, primarily including the study of gamete quality.
Discusssion
Unexplained infertility is diagnosed in couples in the absence of obvious defects of reproductive system. Previously, the signs of UI were clarified: normal fertilization rate, low blastulation rate due to a high frequency of embryo arrest within 3 days of culture, and at the same time, a higher quality of the resulted blastocysts. That is, conception rate was normal in these couples. Most often, embryos stopped developing within 3 days, blastocysts were formed less often, but were of good quality. Therefore, pregnancy can occur, but it takes longer time to conceive. At the same time, the age and increasing comorbidity of patients with UI lead to additional limitation of their reproductive function realization. IVF reduces the time to obtain good-quality blastocysts and, accordingly, the onset of pregnancy, that is, IVF is a method of pathogenetic treatment of UI [5].
The identified impairments of early embryogenesis in patients with UI necessitated the scientific search for possible causes and factors for impairments of early stages of embryogenesis, primarily the genetic ones, which are most conservative, in patients with UI.
Oocyte is a major factor determining human embryonic development [25]. The quality of oocyte is the ability of oocyte to be successfully fertilized and ensures normal embryo development at early stages. The existing evidence does not give grounds for the conclusion that the cause of UI is poor-quality oocytes with a satisfactory number of mature oocytes – more than 6 per puncture (on average 10.8 in our study), satisfactory fertilization rate (76.5 % in our study), possibility of cryopreservation of embryos in 30% of IVF attempts (68.1% in our study) and the frequency of early pregnancy losses comparable to the general population (9.2% in medical history of patients in our study) [5, 26]. At the same time, poor oocyte quality as the cause of UI cannot be ruled out [27].
According to the published data, poor oocyte quality in infertility may be due to gene variants [11, 12]. We carried out a pilot study – whole exome sequencing in 6 married couples (women and men) and purposefully selected them in the group of patients with unexplained infertility to rule out the supposed genetic causes of infertility in married couples with UI.
Whole exome sequencing in the most typical patients with UI found no pathogenic/likely pathogenic variants that could be associated with infertility. In woman in married couple No. 1 and in man in married couple No. 5 the variants of unclear clinical significance were found: FSHR (2-48963163-A-G) and GNRHR (4-67740682-C-T), respectively. It is highly unlikely that these variants could be associated with UI. According to the published data, variant in the FSHR gene (2-48963163-A-G) was described in patient with ovarian dysgenesis and ovarian hyperstimulation syndrome. However, these diseases related to exclusion criteria in the group of UI. Variant in the GNRHR gene (4-67740682-C-T) was described in patients with hypogonadotropic hypogonadism, that also was ruled out, since all men in couples with UI had normozoospermia at the time of inclusion in the study and underwent diagnostic examination by urologist, and were not diagnosed with the disease.
At current stage of the development of science and pharmacology, genetic findings can only influence the decision of patients to use donor gametes in IVF programs. Therefore, the major tactics for management of patients with UI remain unchanged and include performance of IVF as early as possible with good quality embryo transfer (>3, AA, AB, BA according to Gardner grading system) on day 5–6 of culture [5].
Conclusion
A pilot study of whole exome sequencing was performed in 6 married couples (women and men), who were purposefully selected on the basis of clinical and anamnestic findings, laboratory and instrumental parameters, and the results of using assisted reproductive technologies. The known variants that could be associated with infertility were not found in these patients.
Further research is needed to investigate the molecular mechanisms of impairment in early stages of embryogenesis, primarily including the study of gamete quality in patients with unexplained infertility.
References
1. Practice Committee of the American Society for Reproductive Medicine.Electronic address: asrm@asrm.org. Evidence-based treatments for coupleswith unexplained infertility: a guideline. Fertil. Steril. 2020; 113(2): 305-22.https://dx.doi.org/10.1016/j.fertnstert.2019.10.014.
2. ACOG Committee. Infertility workup for the women’s health specialist: ACOGCommittee Opinion, Number 781. Obstet Gynecol. 2019; 133(6): e377-e384.https://dx.doi.org/10.1097/AOG.0000000000003271.
3. Киракосян Е.В., Назаренко Т.А., Бачурин А.В., Павлович С.В. Клиническаяхарактеристика и эмбриологические показатели программ экстракорпорального оплодотворения у женщин с бесплодием неясногогенеза. Акушерство и гинекология. 2022; 5: 83-90. https://dx.doi.org/10.18565/aig.2022.5.83-90. (Kirakosyan E.V., Nazarenko T.A., Bachurin A.V.,
Pavlovich S.V. Clinical characteristics and embryological parametersin IVF programs for women with unexplained infertility. Akusherstvoi Ginekologiya/Obstetrics and Gynecology. 2022; 5: 83-90. (in Russian)).https://dx.doi.org/10.18565/aig.2022.5.83-90.
4. Duffy J.M.N., Adamson G.D., Benson E., Bhattacharya S., Bofill M., Brian K.et al. Top 10 priorities for future infertility research: an internationalconsensus development study. Hum. Reprod. 2020; 35(12): 2715-24.https://dx.doi.org/10.1093/humrep/deaa242.
5. Бачурин А.В., Киракосян Е.В., Назаренко Т.А., Павлович С.В. Анализ эмбриологического этапа программ экстракорпорального оплодотворения упациентов с бесплодием неясного генеза. Акушерство и гинекология.2022; 9: 81-6. https://dx.doi.org/10.18565/aig.2022.9.81-86. (Bachurin A.V.,Kirakosyan E.V., Nazarenko T.A., Pavlovich S.V. Analysis of the embryonicstage of in vitro fertilization programs in patients with unexplained infertility.Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2022; 9: 81-6.(in Russian)). https://dx.doi.org/10.18565/aig.2022.9.81-86
6. Sfakianoudis K., Maziotis E., Karantzali E., Kokkini G., Grigoriadis S., Pantou A.et al. Molecular drivers of developmental arrest in the human preimplantationembryo: a systematic review and critical analysis leading to mapping futureresearch. Int. J. Mol. Sci. 2021; 22(15): 8353. https://dx.doi.org/10.3390/ijms22158353.
7. Jelin A.C., Vora N. Whole exome sequencing: applications in prenatalgenetics. Obstet. Gynecol. Clin. North Am. 2018; 45(1): 69-81. https://dx.doi.org/10.1016/j.ogc.2017.10.003.
8. Salfati E.L., Spencer E.G., Topol S.E., Muse E.D., Rueda M., Lucas J.R. et al.Re-analysis of whole-exome sequencing data uncovers novel diagnostic variantsand improves molecular diagnostic yields for sudden death and idiopathic diseases.Genome Med. 2019; 11(1): 83. https://dx.doi.org/10.1186/s13073-019-0702-2.
9. Суспицын Е.Н., Тюрин В.И., Имянитов Е.Н., Соколенко А.П.Полноэкзомное секвенирование: принципы и диагностические возможности. Педиатр. 2016; 7(4): 142-6. https://dx.doi.org/10.17816/PED74142-146. (Suspitsyn E.N., Tyurin V.I., Imyanitov E.N., Sokolenko A.P.Whole-exome sequencing: principles and diagnostic capabilities. Pediatrician.2016; 7(4): 142-6. (in Russian)). https://dx.doi.org/10.17816/PED74142-146.
10. Gorcenco S., Ilinca A., Almasoudi W., Kafantari E., Lindgren A.G., Puschmann A.New generation genetic testing entering the clinic. Parkinsonism Relat. Disord.2020; 73: 72-84. https://dx.doi.org/10.1016/j.parkreldis.2020.02.015.
11. Chen B., Zhang Z., Sun X., Kuang Y., Mao X., Wang X. et al. Biallelicmutations in PATL2 cause female infertility characterized by oocyte maturationarrest. Am. J. Hum. Genet. 2017; 101(4): 609-15. https://dx.doi.org/10.1016/j.ajhg.2017.08.018.
12. Sang Q., Li B., Kuang Y., Wang X., Zhang Z., Chen B. et al. Homozygous Mutationsin WEE2 Cause Fertilization Failure and Female Infertility. Am. J. Hum. Genet.2018; 102(4): 649-57. https://dx.doi.org/10.1016/j.ajhg.2018.02.015.
13. Miller D.T., Lee K., Chung W.K., Gordon A.S., Herman G.E., Klein T.E. etal. ACMG SF v3.0 list for reporting of secondary findings in clinical exomeand genome sequencing: a policy statement of the American College ofMedical Genetics and Genomics (ACMG). Genet. Med. 2021; 23(8): 1381-90.https://dx.doi.org/10.1038/s41436-021-01172-3.
14. Miller D.T., Lee K., Chung W.K., Gordon A.S., Herman G.E., Klein T.E. et al.Correction to: ACMG SF v3.0 list for reporting of secondary findings in clinicalexome and genome sequencing: a policy statement of the American College ofMedical Genetics and Genomics (ACMG). Genet. Med. 2021; 23(8): 1582-4.https://dx.doi.org/10.1038/s41436-021-01278-8.
15. Рыжкова О.П., Кардымон О.Л., Прохорчук Е.Б., Коновалов Ф.А.,Масленников А.Б., Степанов В.А., Афанасьев А.А., Заклязьминская Е.В.,Ребриков Д.В., Савостьянов К.В., Глотов А.С., Костарева А.А., Павлов А.Е.,Голубенко М.В., Поляков А.В., Куцев С.И. Руководство по интерпретации данных последовательности ДНК человека, полученных методами массового параллельного секвенирования (MPS) (редакция 2018,версия 2). Медицинская генетика. 2019; 18(2): 3-23. https://dx.doi.org/10.25557/2073-7998.2019.02.3-23. (Ryzhkova O.P., Kardymon O.L.,Prohorchuk E.B., Konovalov F.A., Maslennikov A.B., Stepanov V.A.,Afanasyev A.A., Zaklyazminskaya E.V., Rebrikov D.V., Savostianov K.V.,Glotov A.S., Kostareva A.A., Pavlov A.E., Golubenko M.V., Polyakov A.V.,Kutsev S.I. Guidelines for the interpretation of massive parallel sequencingvariants (update 2018, v2). Medical genetics. 2019; 18(2): 3-24 (in Russian)).https://dx.doi.org/10.25557/2073-7998.2019.02.3-23.
16. Rehm H.L., Bale S.J., Bayrak-Toydemir P., Berg J.S., Brown K.K., Deignan J.L. etal. ACMG clinical laboratory standards for next-generation sequencing. Genet.Med. 2013; 15(9): 733-47. https://dx.doi.org/10.1038/gim.2013.92.
17. Ensembl Variant Effect Predictor (VEP). Available at: https://www.ensembl.org/info/docs/tools/vep/index.html
18. gnomAD - Genome Aggregation Database. Available at: https://gnomad.broadinstitute.org/
19. OMIM. An Online Catalog of Human Genes and Genetic Disorders. Availableat: https://omim.org/
20. ClinVar. Available at: https://www.ncbi.nlm.nih.gov/clinvar/
21. LOVD v.3.0 - Leiden Open Variation Database. Online gene-centered collectionand display of DNA variants. Available at: https://www.lovd.nl/
22. RUSeq - проект по объединению генетической информации между клиническими лабораториями и геномными центрами России. Доступнопо: https://ruseq.ru/#/ (RUSeq - a project to pool genetic informationbetween clinical laboratories and genomic centers in Russia. Available at:https://ruseq.ru/#/ (in Russian)).
23. Barbitoff Y.A., Khmelkova D.N., Pomerantseva E.A., Slepchenkov A.V.,Zubashenko N.A., Mironova I.V. et al. Expanding the Russian allele frequencyreference via cross-laboratory data integration: insights from 6,096 exomesamples. medRxiv. 2021. https://dx.doi.org/10.1101/2021.11.02.21265801.
24. Human Phenotype Ontology (HPO). Available at: https://hpo.jax.org/app/
25. Larbuisson A., Raick D., Demelenne S., Delvigne A. ICSI diagnostic: a way toprevent total fertilization failure after 4 unsuccessful IUI. Basic Clin. Androl.2017; 27: 18. https://dx.doi.org/10.1186/s12610-017-0061-z.
26. Bosselut H., Paulmyer-Lacroix O., Gnisci A., Bretelle F., Perrin J., Courbiere B.Facteurs pronostiques des chances de naissance vivante enfecondation in vitropour infertilite inexpliquee: etude de cohorte. (Prognostic factors of live-birth afterin vitro fertilization for unexplained infertility: A cohort study). Gynecol. Obstet.Fertil. Senol. 2021; 49(7-8): 601-7. (in French). https://dx.doi.org/10.1016/j.gofs.2021.01.002.
27. Киракосян Е.В., Екимов А.Н., Павлович С.В. Значение ооцитарногофактора в развитии бесплодия неясного генеза. Акушерство и гинекология. 2022; 1: 14-21. https://dx.doi.org/10.18565/aig.2022.1.14-21.(Kirakosyan E.V., Ekimov A.N., Pavlovich S.V. The significance of theoocyte factor in the development of infertility of unclear genesis. Akusherstvoi Ginekologiya/Obstetrics and Gynecology. 2022; 1: 14-21. (in Russian)).https://dx.doi.org/10.18565/aig.2022.1.14-21.
Received 17.10.2022
Accepted 28.11.2022
About the Authors
Evgeniya V. Kirakosyan, Ph.D. student, Department of Obstetrics, Gynecology, Perinatology and Reproductology, I.M. Sechenov First MSMU, Ministry of Health of Russia (Sechenov University), Academician V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia, +7(916)574-79-63, evgeniya.kirakosyan@mail.ru,https://orcid.org/0000-0002-6021-2449, 4 Akademika Oparina str., Moscow 117997, Russia.
Ekaterina A. Pomerantseva, Ph.D., geneticist, Department of Clinical Genetics, Institute of Reproductive Genetics, Academician V.I. Kulakov NMRC for OG&P,
Ministry of Health of Russia, e.pomerantseva@gmail.com, https://orcid.org/0000-0002-2027-2738, 4 Akademika Oparina str., Moscow 117997, Russia.
Stanislav V. Pavlovich, Ph.D., Academic Secretary, Academician V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia; Professor, Department of Obstetrics, Gynecology, Perinatology and Reproductology, I.M. Sechenov First MSMU, Ministry of Health of Russia (Sechenov University), +7(495)438-20-88, s_pavlovich@oparina4.ru,
https://orcid.org/0000-0002-1313-7079, 4 Akademika Oparina str., Moscow 117997, Russia.
Corresponding author: Evgeniya V. Kirakosyan, evgeniya.kirakosyan@mail.ru
Authors’ contributions: Kirakosyan E.V., Pomerantseva E.A., Pavlovich S.V. – the concept and design of the study, analysis of published literature, article editing; Kirakosyan E.V., Pomerantseva E.A. – material collection and processing; statistical data processing; Kirakosyan E.V. – article writing.
Conflicts of interest: The authors declare that they have no conflicts of interest.
Funding: The study was funded by the Laboratory of the Institute of Reproductive Genetics of Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia.
Ethical Approval: The protocol of the study was approved by the Academic Council of I.M. Sechenov First Moscow State Medical University (Sechenov University), Ministry of Health of Russia (Extract from Order No. 4070/OP-32 dated 30.09.2020), the local Ethics Committee of Sechenov University, Ministry of Health of Russia (Extract from Protocol No. 33-20, dated 25.11.2020), the Commission on the Ethics of Biomedical Research of Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia (Extract from Protocol No. 11 dated 12.11.2020).
Patients’ Consent for Publication: The patients have signed informed consent for participation in the study, including the use of their data in publications.
Authors' Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator.
For citation: Kirakosyan E.V., Pomerantseva E.A., Pavlovich S.V. Whole exome sequencing in couples with unexplained infertility (pilot study).
Akusherstvo i Gynecologia/Obstetrics and Gynecology. 2022; 12: 115-121 (in Russian)
https://dx.doi.org/10.18565/aig.2022.247



