Differential gene expression in uterine fibroids
Kuznetsova M.V., Nersesyan E.A., Burmenskaya O.V., Tonoyan N.M., Mikhailovskaya G.V., Svirepova K.A., Adamyan L.V., Trofimov D.Yu.
Objective: The study of differential gene expression in fibroid nodules and myometrium to identify potential target genes for the development of personalized approaches to the diagnosis, prediction of the course, prevention and treatment of uterine fibroids (also known as leiomyomas).
Materials and methods: The total number of 133 tissue samples were examined, including 48 samples of myometrium and 85 samples of fibroid nodules. The material was obtained during surgical interventions.
All samples of fibroid nodules were analyzed for the presence of somatic mutations in exon 2 of the MED12+ gene. In RNA samples, which were isolated from myometrial tissues and fibroid nodules using RT-PCR, gene expression analysis of the HMGA2, PAPPA1, GRPR, TYMS, PLAG1, VCAN, AVPR1A, ESR1, PLA2R1, RANKL, KLF11, KRT19, MMP11, ADAM12, MMP16, PCP4, STAB2, WIF1 genes was performed.
Results: Somatic mutations in the MED12 gene were found in 42 samples (MED 12+ fibroid nodules), and no mutations were found in 43 samples (MED12- fibroid nodules). Multiple fibroids were found in 19 patients among the studied patient sample, while fibroid nodules both MED12+ and MED12- were found in 9 patients. The results of our study showed statistically significant differences in gene expression levels between myometrium and fibroid nodules in 13 of 18 examined genes. At the same time, a number of genes demonstrated opposite changes in the transcription level compared to myometrium, depending on the presence of somatic mutation
in the MED12 gene.
It has been shown that in the presence of somatic mutation in the MED12 gene, significantly increased gene expression of 7 genes – GRPR, TYMS, RANKL, MMP11, AVPR1A, PCP4 and ESR1 and significantly reduced gene expression of 2 genes – KRT19, KLF11 was in nodules compared to myometrium. There was significantly increased expression of only 2 genes – GRPR and TYMS in all fibroid nodules, whereas increased expression of the PAPPA1, VCAN, ESR1, RANKL, KRT19, MMP11, PCP4 and AVPR1A genes in MED12+ nodule phenotype was found compared to MED12- nodules.
Conclusion: The obtained data enabled to distinguish two molecular phenotypes of uterine fibroids in the examined sample – the associated and unassociated phenotype with somatic mutation in exon 2 of the MED12 gene. The expression of some genes alters in different molecular phenotypes of uterine fibroids, and statistically significant differences are found not only between each type of uterine fibroids and myometrium, but also between different molecular phenotypes of fibroids. The obtained results enable to consider some of the examined genes as potential targets to develop personalized approaches to diagnosis, prognosis of the course, prevention and treatment of uterine fibroids.
Authors' contributions: Nersesyan E.A., Kuznetsova M.V., Adamyan L.V. – the concept and design of the study; Nersesyan E.A., Kuznetsova M.V., Burmenskaya O.V., Tonoyan N.M., Mikhailovskayа G.V., Svirepova K.A. – material collection and processing; Nersesyan E.A. – statistical data processing; Kuznetsova M.V., Nersesyan E.A. – text writing; Trofimov D.Yu., Adamyan L.V. – text editing.
Conflicts of interest: The authors confirm that they have no conflict of interest to declare.
Funding: Project of the Russian Science Foundation No. 23-15-00069 “Development of preventive vaccine against leiomyoma in patients planning pregnancy”.
Ethical Approval: The study was approved by Ethics Committee of the Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia.
Patient Consent for Publication: All patients have signed voluntary informed consent for publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator.
For citation: Kuznetsova M.V., Nersesyan E.A., Burmenskaya O.V., Tonoyan N.M., Mikhailovskaya G.V., Svirepova K.A., Adamyan L.V., Trofimov D.Yu.
Differential gene expression in uterine fibroids.
Akusherstvo i Gynecologia/Obstetrics and Gynecology. 2024; (8): 87-95 (in Russian)
https://dx.doi.org/10.18565/aig.2024.72
Keywords
Uterine fibroids are an important medical and social problem due to high prevalence, unclear etiopathogenesis, lack of algorithms for optimal treatment and prevention of this condition. In the general structure of gynecological pathologies, uterine fibroids rank second after pelvic inflammatory disease. Uterine fibroids are diagnosed in 25–75% of reproductive-age women, 5–10% of them have uterine factor infertility [1–3].
Myomectomy is the most effective treatment of symptomatic uterine fibroids in women of reproductive age, who are planning pregnancy. However, it is an approach that indicates high probability of recurrent disease almost in 90% of patients. Repeated surgeries are performed in 27% of patients, who previously underwent surgery for a single uterine fibroid, and 59% of patients had surgery for multiple uterine fibroids [1, 2, 4–7].
Radical hysterectomy is an option to prevent recurrence of leiomyoma. However, this option is unacceptable for women, who are planning pregnancy. Moreover, a significant problem is that myomectomy causes trauma to the uterus, scar formation leading to impaired reproductive performance during pregnancy and the risk of uterine ruptures during stretch and contractions before and after childbirth. In addition, maximum uterine fibroids growth rate is during pregnancy and is associated with high progesterone levels in the blood [8]. Thus, one of the most important medical and social issues of leiomyoma treatment is development of treatment methods to prevent disease recurrence in patients, who are planning pregnancy or are childbearing women. Therefore, systematization of existing methods to prevent recurrence of leiomyomas or to slow the growth of existing fibroid nodules in patients, who make plans for childbirth, or in pregnant patients is highly relevant.
Uterine fibroids are monoclonal benign tumor originating from myocytes of the cervix and/or the body of the uterus [1]. It is believed that each fibroid nodule is starts to grow in a single primary cell with genetic damage called as “driver mutation” [9]. These mutations may include rearrangements involving 6p21 and 10q22 loci, deletions affecting 1p and 3q loci [9, 10, 11], as well as somatic mutations in the MED12 gene leading to changes in expression of the HGMA1, HGMA2, FH, BHD, TSC2, PCOLCE, ORC5L, LHFPL3, ESR2 and RAD51 genes in tissues of fibroid nodules [12]. It was found that multiple fibroid nodules in the same uterus can have different genetic defects, that indicates again that each fibroid nodule develops independently [10, 13].
Most significant somatic mutations are MED12 mutations, that are identified in 50–70% patients with uterine fibroids. Most often, these are missense mutations in codons 43 and 44, as well as deletions of different length. The MED12 gene is located on the X chromosome and encodes a 250 kDa protein, that is a subunit of the large mediator complex involved in transcription regulation of the RNA polymerase II complex. The role of MED12 mutations in the development of leiomyomas remains to be clarified. It is assumed that these mutations can change the functional properties of the MED12 protein, and this mutant protein modifies the functions of the RNA polymerase II complex in the cells of growing leiomyoma, leading to changes in the activity of other genes encoding proteins, which are involved in tumor growth and progression [14, 15].
It has been shown that the cells in fibroid nodules have significant epigenetic changes in comparison to the tissues of morphologically unchanged myometrium. The changes in the promoter methylation levels and accompanying changes in mRNA expression in 55 genes were detected in uterine fibroid cells [16]. In addition, the differences in methylation levels and RNA expression in 120 genes were identified between leiomyoma cells and normal cells of the morphologically unchanged myometrium [17].
In general, the studies on the differences in expression profile in myometrium and uterine fibroids of different molecular phenotypes lead to further development and improvement of approaches to individualize the prognosis for the course and treatment of uterine fibroids in patients. In addition, this kind of studies identify potential target genes to develop new methods of therapeutic impact on tumors and prevention of disease development.
The purpose of the study was differential gene expression analysis in fibroid nodules and myometrium as the basis for further development of personalized approaches to the diagnosis, prediction of the course and treatment of uterine fibroids (also known as leiomyomas).
Materials and methods
The study material is represented by total number of 133 tissue samples of uterine fibroids (85 samples) and morphologically unchanged myometrium (48 samples), which were obtained from 48 women, who underwent surgery for uterine fibroids at the Department of Operative Gynecology, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia (Kulakov Center) as well as at gynecological departments of the Central Clinical Hospital with Polyclinic of the Administrative Directorate of the President of the Russian Federation and City Clinical Hospital of Healthcare Department of Moscow in 2022–2023.
Molecular genetic testing was performed in the Laboratory of Molecular Genetic Methods and the Laboratory of Oncological Genetics, the Institute of Reproductive Genetics of Kulakov Center.
Based on bioinformatic analysis, 18 candidate genes were selected (HMGA2, PAPPA1, GRPR, TYMS, PLAG1, VCAN, AVPR1A, ESR1, PLA2R1, RANKL, KLF11, KRT19, MMP11, ADAM12, MMP16, PCP4, SATB2, WIF1), that are presumably involved in pathogenesis of uterine fibroids [18–20].
The samples of each uterine fibroid, that was removed in patients, were analyzed for the presence of somatic mutations in exon 2 of the MED12 gene (MED12 +) using Sanger sequencing. In addition, sequencing of exon 2 of the MED12 gene was performed in peripheral blood samples collected from all patients to confirm the somatic nature of mutations.
Expression levels of the HMGA2, PAPPA1, GRPR, TYMS, PLAG1, VCAN, AVPR1A, ESR1, PLA2R1, RANKL, KLF11, KRT19, MMP1, ADAM12, MMP16, PCP4, SATB2, WIF1 genes in tissue samples of fibroid nodules and morphologically unchanged myometrium was analyzed using reverse transcription polymerase chain reaction. Two repeats of amplification were performed for each marker. The mean value of Ct was calculated according to the method developed by Burmenskaya O.V. et al. [21]. After amplification the level of transcript representation was estimated using comparative ΔCt method. Normalization factor was calculated using reference genes B2M, GUSB [22]. The median value (Me) in the comparison group was taken as 1, and the expression indices in the samples were calculated with regard to this value.
Statistical analysis
IBM SPSS Statistics 17.0. was used for statistical analysis. The differences were considered to be statistically significant at p<0.05. Median (Me) was calculated as a measure of central tendency in the study groups. The obtained data in the groups are represented as median (Me) and the first and third interquartiles (Q1, Q3).
Results and discussion
Out of 48 patients in the study sample, 29 women underwent surgical treatment for a single uterine fibroid, and 19 women for multiple uterine fibroids.
There were no mutations in the MED12 gene detected in peripheral blood samples obtained from 48 patients, that indicate somatic origin of these mutations in precursor cells of uterine fibroids.
Table 1 presents the results of mutation testing in the MED12 gene in specimens of fibroid nodules obtained from 48 patients in the study sample.
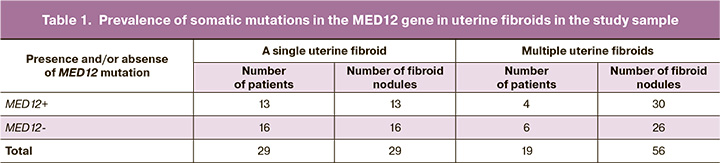
Somatic mutations in the MED12 gene were found in 13 patients, who underwent surgery for a single uterine fibroid. In 16 patients who underwent surgery for a single uterine fibroid, no somatic mutations in the MED12 gene were detected in the samples of fibroid nodules. In 4 patients, who had surgery for multiple uterine fibroids, MED12 mutations were found in all 30 fibroid nodules. In 6 patients, no MED12 mutations were present in 26 uterine fibroid nodules. Moreover, in 9 patients with multiple uterine fibroids, there were both MED12 mutations and no mutations detected in 29 fibroid nodules.
Table 2 represents the results of comparative gene expression analysis in 18 genes in the specimens of myometrium and uterine fibroids in the study sample in general.
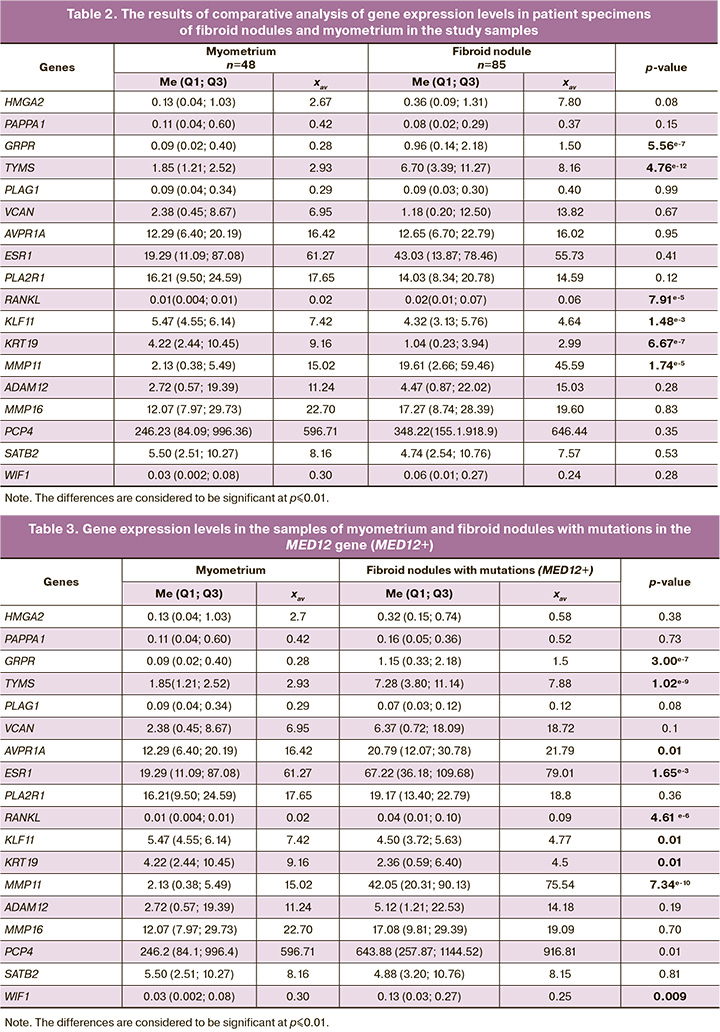
Significanly high levels of expression of the GRPR, TYMS, RANKL and MMP11 genes and low levels of expression of the KLF11 and KRT19 genes were in the total number of fibroid nodules samples compared with myometrium. The obtained data indicate that the GRPR, TYMS, RANKL, MMP11 genes are involved in the pathogenesis of uterine fibroids despite the presence or absence of somatic mutations in the MED12 gene. In addition, there is a significant difference in the median values in expression of the HMGA2 gene, which is known as one of the key genes, and overexpression of this gene leads to development of uterine fibroids [21]. However, p-value for this gene does not demonstrate the reliability of the difference. This is primarily due to highly variable data within the group of uterine fibroids, that required division of the general group into subgroups for further analysis.
It is known that the GRPR, TYMS, RANKL и MMP11 genes are often overexpressed in some types of tumors (particularly in breast cancer, lung cancer, melanoma and some other) [23–25]. At the same time, it has been shown that the KLF11 and KRT19 genes demonstrate reduced expression, especially in fibroids mainly due to methylation of the promoter regions of these genes in tumor cells [26].
The next step of data analysis was comparison of obtained data between fibroid nodules with and without MED12 mutations, and myometrium. Table 3 represents expression levels of MED12+ in the examined genes in myometrium and fibroid nodules.
In uterine fibroids with somatic mutations in the MED12 gene, expression of the GRPR, TYMS, RANKL MMP11 genes, which were mentioned in analysis of the total number of nodules sample, was significantly high. However, significantly increased expression of other three genes – AVPR1A, PCP4 and ESR1 was detected. MED12+ nodules are characterized by elevated expression level of estrogen receptor 1 (ESR1) in uterine fibroid cells, that leads to increased response to estrogen and tumor growth. The product of the AVPR1A gene is a vasopressin receptor and is involved in the mechanisms of cell regulation and cell proliferation of some tumors (in particular, thyroid carcinoma) [27].
As is shown in Table 3, in fibroid nodules with somatic mutations in the MED12 gene, statistically significant reduction in expression was not only in the KRT19, KLF11 genes, but also in the WIF1 gene compared with myometrium. The WIF1 gene is a well-known tumor suppressor, which is switched off in cells during diverse oncogenic processes, and reduction of its activity is also achieved through the mechanism of methylation in promoter region [28].
Table 4 represents candidate gene expression levels in the samples of myometrium and the second subgroups of myomas – fibroid nodules without mutations in the MED12 gene.
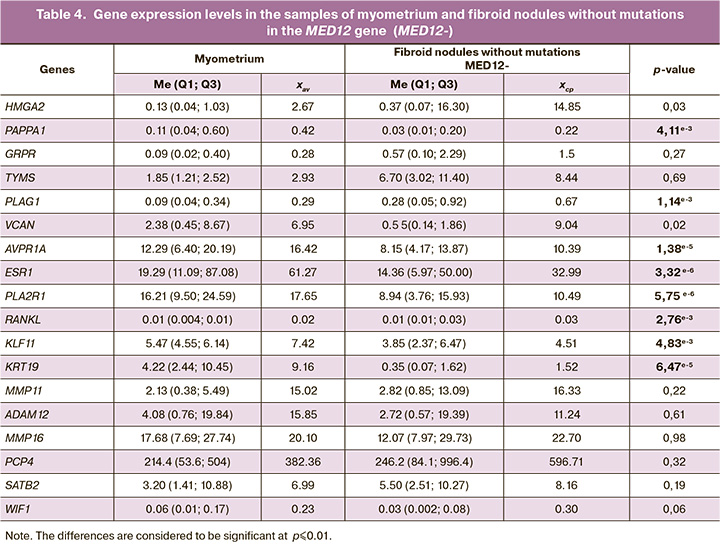
In this sample we observe another picture of reduction/increase in gene expression versus the MED12+ nodules. Elevated expression of the PLAG1 and RANKL genes in this subgroup of nodules was found compared with myometrium, while expression levels of the PAPPA1, ESR1, PLA2R1, KRT19, KLF11 and AVPR1A genes were reduced compared with myometrium. That is, the result for the ESR1, AVPR1A genes showed opposite values compared to the values obtained for MED12+. In different molecular subtypes of nodules, we observe opposite changes in expression of these genes.
To identify further differences between the two molecular phenotypes of uterine fibroids, comparative analysis of expression of all these genes in fibroid nodule samples with and without somatic MED12 mutations was performed. The results of comparative analysis are represented in Table 5.
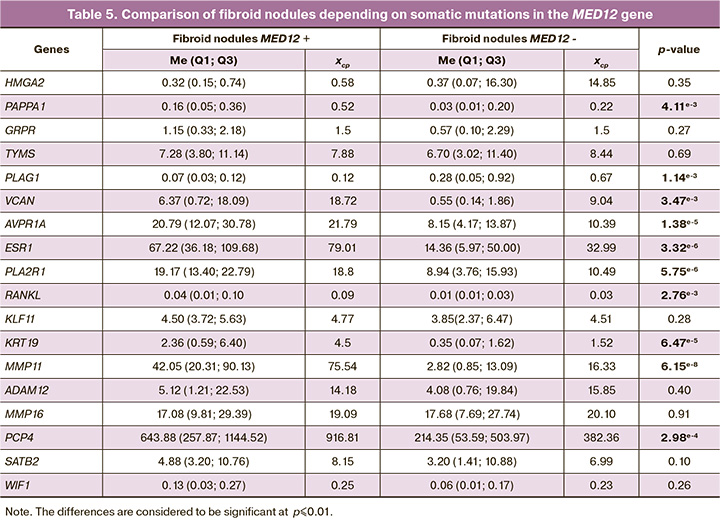
The spectrum of differences between the two subgroups of fibroid nodules was higher than between each of these subtypes and myometrium. In total, statistically significant differences were found for 10 genes.
For the PAPPA1, VCAN, ESR1, RANKL, KRT19, MMP11, PCP4 and AVPR1A genes elevated expression was found in MED12+ phenotype of nodules compared with the MED12- nodules. The values of three genes – PLAG1, WIF1, KLF11 in one type of nodules were not significantly different from the values in myometrium, while in the other type of nodules were different. Reduced expression of KRT19, KLF11 was detected both in total number of fibroid nodules sample and in the subgroups. At the same time, comparison between the subgroups of nodules showed that in the subgroup MED12+, expression level of KRT19 was significantly higher compared with MED12-. There was significantly elevated expression of the GRPR and TYMS genes in all fibroid nodules, but no difference was found between the two phenotypes of nodules. Significantly elevated expression of the VCAN gene was not detected in the MED12- nodules, although the mean value differred significantly. At the same time, in the MED12+ nodules, the mean values and p-values are higher, that can be explained by data variability within the groups. A similar situation applies to the HMGA2 gene due to highly variable data of gene expression. At the same time, no statistically significant differences were detected. Hyperexpression of HMGA2 in uterine fibroids was found quite a long time ago and is considered to be a key point in the genesis of leiomyomas, where chromosomal rearrangements occur involving 12q14-15 [29]. However, previously published data on gene expression related to differences between uterine leiomyomas with and without 12q14-15 aberrations, are not enough to understand how often this gene is overexpressed in fibroids in general. The data obtained by us complement the existing picture: in the study sample, no hyperexpression of the HMGA2 gene in the MED12+ nodules was detected, whereas in the group of the MED12- nodules, high expression levels of HMGA2 were found in 17/43 (40%) nodules. Therefore, among the MED12- nodules, there is another subgroup of tumors characterized by high expression of the HMGA2 gene, that requires more thorough study.
It should be noted that most genes – HMGA2, PAPPA1, GRPR, TYMS, PLAG1, VCAN, AVPR1A, ESR1, PLA2R1, RANKL, KLF11, KRT19, MMP11, PCP4, SATB2, WIF1, which were primarily selected as potential pathogenetic markers of uterine fibroids, demonstrated differential expression in myomas, that is, they are somehow involved in the process of tumor development.
Conclusion
The results obtained in analysis of differential expression of 18 genes in uterine fibroids and myometrium enable to distinguish two molecular phenotypes of uterine fibroids – the associated (MED12+) and unassociated (MED12-) phenotypes with somatic mutation in exon 2 of the MED12 gene.
Molecular phenotypes of uterine fibroids associated and unassociated with somatic mutations in the MED 12 gene, occur both in single and multiple uterine fibroids. Our study showed that expression of some genes changes in different molecular phenotypes of uterine fibroids, and statistically significant differences are found not only between each type of fibroids and myometrium, but also between different molecular phenotypes of fibroids.
All of the above makes it possible to formulate a hypothesis that different molecular phenotypes of uterine fibroids can differ in both gene expression profiles of tumor markers and different potential for the growth and recurrence, that requires further research. The obtained results enable to consider some of the examined genes as potential targets to develop personalized approaches to diagnosis, prognosis of the course, prevention and treatment of uterine fibroids.
References
- Адамян Л.В., Андреева Е.Н., Артымук Н.В., Белоцерковцева Л.Д., Беженарь В.Ф., Геворкян М.А., Глухов Е.Ю., Гус А.И., Доброхотова Ю.Э., Жорданиа К.И., Зайратьянц О.В., Козаченко А.В., Киселев С.И., Коган Е.А., Кузнецова И.В., Курашвили Ю.Б., Леваков С.А., Малышкина А.И., Мальцева Л.И., Марченко Л.А., Мурватов К.Д., Пестрикова Т.Ю., Попов А.А., Протопопова Н.В., Самойлова А.В., Сонова М.М., Тихомиров А.Л., Ткаченко Л.В., Урумова Л.Т., Филиппов О.С., Хашукоева А.З., Чернуха Г.Е., Ярмолинская М.И., Яроцкая Е.Л. Миома матки: диагностика, лечение и реабилитация. Клинические рекомендации (протокол лечения). М.; 2015. [Adamyan L.V., Andreeva E.N., Artymuk N.V., Belotserkovtseva L.D., Bezhenar V.F., Gevorkyan M.A., Glukhov E.Yu., Gus A.I., Dobrokhotova Y.E., Zhordania K.I., Zayratyants O.V., Kozachenko A.V., Kiselev S.I., Kogan E.A., Kuznetsova I.V., Kurashvili Yu.B., Levakov S.A., Malyshkina A.I., Maltseva L.I., Marchenko L.A., Murvatov K.D., Pestrikova T.Yu., Popov A.A., Protopopova N.V., Samoilova A.V., Sonova M.M., Tikhomirov A.L., Tkachenko L.V., Urumova L.T., Filippov O.S., Khashukoeva A.Z., Chernukha G.E., Yarmolinskaya M.I., Yarotskaya E.L. Uterine myoma: diagnosis, treatment and rehabilitation. Clinical guidelines (treatment protocol). Moscow; 2015. (in Russian)].
- Савельева Г.М., Сухих Г.Т., Серов В.Н., Радзинский В.Е., Манухин И.Б., ред. Гинекология. Национальное руководство. 2-е изд. М.; ГЭОТАР-Медиа; 2020. 1044 с. [Savelyeva G.M., Sukhikh G.T., Serov V.N., Radzinsky V.E., Manukhin I.B., eds. Gynecology. National Guide. 2nd ed. Moscow; GEOTAR-Media; 2020. 1044 p. (in Russian)].
- Stewart E.A., Cookson C.L., Gandolfo R.A., Schulze-Rath R. Epidemiology of uterine fibroids: a systematic review. BJOG. 2017; 124(10): 1501-12. https://dx.doi.org/10.1111/1471-0528.14640.
- Тихомиров А.Л., Лубнин Д.М. Миома матки. М.: ООО Медицинское информационное агентство; 2006. 176 с. [Tikhomirov A.L., Lubnin D.M. Uterine myoma. M.: Medical News Agency LLC; 2006. 176 p. (in Russian)].
- Беженарь В.Ф., Комличенко Э.В., Ярмолинская М.И., Дедуль А.Г., Шевелева Т.С., Малушко А.В., Калинина Е.А., Зубарева Т.М., Гамзатова З.Х., Кондратьев А.А. Инновационные подходы к восстановлению репродуктивной функции у больных с миомой матки. Акушерство и гинекология. 2016; 1: 80-7. [Bezhenar V.F., Komlichenko E.V., Yarmolinskaya M.I., Dedul A.G., Sheveleva T.S., Malushko A.V., Kalinina E.A., Zubareva T.M., Gamzatova Z.Kh., Kondratyev A.A. Innovative approaches to reproductive function recovery in patients with uterine myoma. Obstetrics and Gynecology. 2016; (1): 80-7. (in Russian)]. https://dx.doi.org/10.18565/aig.2016.1.80-87.
- Серов В.Н., Сухих Г.Т., ред. Клинические рекомендации. Акушерство и гинекология. 4-е изд. М: ГЭОТАР-Медиа; 2014. 1024 с. [Serov V.N., Sukhikh G.T., eds. Clinical guidelines. Obstetrics and gynecology. 4th ed. Moscow: GEOTAR-Media; 2014. 1024 p. (in Russian)].
- Vidal-Mazo C., Forero-Diaz C., Lopez-Gonzalez E., Yera-Gilabert M., Machancoses F.H. Clinical recurrence of submucosal myoma after a mechanical hysteroscopic myomectomy: Review after 5 years follow up. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019; 243: 41-5. https://dx.doi.org/10.1016/j.ejogrb.2019.10.014.
- Marugo M., Centonze M., Bernasconi D., Fazzuoli L., Berta S., Giordano G. Estrogen and progesterone receptors in uterine leiomyomas. Acta Obstet. Gynecol. Scand. 1989; 68(8): 731-5. https://dx.doi.org/10.3109/00016348909006147.
- Wise L.A., Laughlin-Tommaso S.K. Epidemiology of uterine fibroids – from menarche to menopause. Clin. Obstet. Gynecol. 2016; 59(1): 2-24. https://dx.doi.org/10.1097/GRF.0000000000000164.
- Sparic R., Mirkovic L., Malvasi A., Tinelli A. Epidemiology of uterine myomas: a review. Int. J. Fertil. Steril. 2016; 9(4): 424-5. https://dx.doi.org/10.22074/ijfs.2015.4599.
- Manta L., Suciu N., Toader O., Purcărea R.M., Constantin A., Popa F. The etiopathogenesis of uterine fibromatosis. J. Med. Life. 2016; 9(1): 39-45.
- McWilliams M.M., Chennathukuzhi V.M. Recent advances in uterine fibroid etiology. Semin. Reprod. Med. 2017; 35(2): 181-9. https://dx.doi.org/10.1055/s-0037-1599090.
- Markowski D.N., Bartnitzke S., Löning T., Drieschner N., Helmke B.M., Bullerdiek J. MED12 mutations in uterine fibroids--their relationship to cytogenetic subgroups. Int. J. Cancer. 2012; 131(7): 1528-36. https://dx.doi.org/10.1002/ijc.27424.
- Narayanan D.L., Phadke S.R. A novel variant in MED12 gene: further delineation of phenotype. Am. J. Med. Genet. A. 2017; 173(8): 2257-60. https://dx.doi.org/10.1002/ajmg.a.38295.
- Mittal P., Shin Y.H., Yatsenko S.A., Castro C.A., Surti U., Rajkovic A. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J. Clin. Invest. 2015; 125(8): 3280-4. https://dx.doi.org/10.1172/JCI81534.
- Navarro A., Yin P., Monsivais D., Lin S.M., Du P., Wei J.-J., Bulun S.E. Genome-wide DNA methylation indicates silencing of tumor suppressor genes in uterine leiomyoma. PLoS One. 2012; 7(3): e33284. https://dx.doi.org/10.1371/journal.pone.0033284.
- Maekawa R., Sato S., Yamagata Y., Asada H., Tamura I., Lee L. et al. Genome-wide DNA methylation analysis reveals a potential mechanism for the pathogenesis and development of uterine leiomyomas. PLoS One. 2013; 8(6): e66632. https://dx.doi.org/10.1371/journal.pone.0066632.
- Maekawa R., Sato S., Tamehisa T., Sakai T., Kajimura T., Sueoka K. et al. Different DNA methylome, transcriptome and histological features in uterine fibroids with and without MED12 mutations. Sci. Rep. 2022; 12(1): 8912. https://dx.doi.org/10.1038/s41598-022-12899-7.
- Mehine M., Kaasinen E., Heinonen H.R., Makinen N., Kampjarvi K., SarvilinnaN. et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc. Natl. Acad. Sci. U. S. A. 2016; 113(5): 1315-20. https://dx.doi.org/10.1073/pnas.1518752113.
- Yin P., Ono M., Moravek M.B., Coon J.S., Navarro A., Monsivais D. et al. Human uterine leiomyoma stem/progenitor cells expressing CD34 and CD49b initiate tumors in vivo. J. Clin. Endocrinol. Metab. 2015; 100(4): E601-6. https://dx.doi.org/10.1210/jc.2014-2134.
- Бурменская О.В., Трофимов Д.Ю., Кометова В.В., Сергеев И.В., Маерле А.В., Родионов В.В., Сухих Г.Т. Разработка и опыт использования транскрипционной сигнатуры генов в диагностике молекулярных подтипов рака молочной железы. Акушерство и гинекология. 2020; 2: 132-40. [Burmenskaya O.V., Trofimov D.Yu., Kometova V.V., Sergeev I.V., Maerle A.V., Rodionov V.V., Sukhikh G.T. Development and experience of using the transcriptional gene signature in the diagnosis of molecular breast cancer subtypes. Obstetrics and Gynecology. 2020; (2): 132-40. (in Russian)]. https://dx.doi.org/10.18565/aig.2020.2.132-140.
- Schmittgen T.D., Zakrajsek B.A., Mills A.G., Gorn V., Singer M.J., Reed M.W. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 2000; 285(2): 194-204. https://dx.doi.org/10.1006/abio.2000.4753.
- Zhang H., Qi L., Cai Y., Gao X. Gastrin-releasing peptide receptor (GRPR) as a novel biomarker and therapeutic target in prostate cancer. Ann. Med. 2024; 56(1): 2320301. https://dx.doi.org/10.1080/07853890.2024.2320301.
- Yunchu Y., Miyanaga A., Matsuda K., Kamio K., Seike M. Exploring effective biomarkers and potential immune related gene in small cell lung cancer. Sci. Rep. 2024; 14(1): 7604. https://dx.doi.org/10.1038/s41598-024-58454-4.
- Schaper-Gerhardt K., Gutzmer R., Angela Y., Zimmer L., Livingstone E., Schadendorf D. et al. The RANKL inhibitor denosumab in combination with dual checkpoint inhibition is associated with increased CXCL-13 serum concentrations. Eur. J. Cancer. 2024; 202: 113984. https://dx.doi.org/10.1016/j.ejca.2024.113984.
- Кузнецова М.В., Тоноян Н.М., Свирепова К.А., Адамян Л.В., Трофимов Д.Ю. Роль метилирования генов в развитии миомы матки. Проблемы репродукции. 2023; 29(1): 33-8. [Kuznetsova M.V., Tonoyan N.M., Svirepova K.A., Adamyan L.V., Trofimov D.Yu. The role of gene methylation in the development of fibroids. Russian Journal of Human Reproduction. 2023; 29(1): 33-8. (in Russian)]. https://dx.doi.org/10.17116/repro20232901133.
- Pan L., Zhang L., Fu J., Shen K., Zhang G. Integrated transcriptome sequencing and weighted gene co-expression network analysis reveals key genes of papillary thyroid carcinomas. Heliyon. 2024; 10(7): e27928. https://dx.doi.org/10.1016/j.heliyon.2024.e27928.
- Sarangi J., Das P., Ahmad A., Sulaiman M., Ghosh S., Gupta B. et al. Methylation study of tumor suppressor genes in human aberrant crypt foci, colorectal carcinomas, and normal colon. J. Cancer Res. Ther. 2024; 20(1): 268-74. https://dx.doi.org/10.4103/jcrt.jcrt_1573_22.
- Klemke M., Meyer A., Nezhad M.H., Bartnitzke S., Drieschner N., Frantzen C. et al. Overexpression of HMGA2 in uterine leiomyomas points to its general role for the pathogenesis of the disease. Genes Chromosomes Cancer. 2009; 48(2): 171-8. https://dx.doi.org/10.1002/gcc.20627.
Received 26.03.2024
Accepted 12.07.2024
About the Authors
Maria V. Kuznetsova, PhD (Bio), Senior Researcher at the Laboratory of Molecular Genetic Methods of the Institute of Reproductive Genetics, V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia, +7(495)438-13-41, mkarja@mail.ru, https://orcid.org/0000-0003-3790-0427Ekaterina A. Nersesyan, PhD student at the Department of Reproductive Medicine and Surgery, A.I. Yevdokimov Moscow State University of Medicine and Dentistry,
Ministry of Health of Russia, +7(916)182-22-40, knea_doc@icloud.com, https://orcid.org/0009-0004-6444-5962
Olga V. Burmenskaya, Dr. Bio. Sci., Head of the Laboratory of Oncological Genetics, V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia, +7(495)438-22-92, o_bourmenskaya@oparina4.ru,
https://orcid.org/0000-0003-2842-3980
Narine M. Tonoyan, PhD, obstetrician-gynecologist at the Department of Operative Gynecology, V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia, https://orcid.org/0000-0002-1631-1829
Galina V. Mikhaylovskaya, Biologist at the Laboratory of Molecular Genetic Methods of the Institute of Reproductive Genetics, V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia, +7(495)438-13-41, galina.mikhaylovskaya@gmail.com
Ksenia A. Svirepova, doctor of clinical laboratory diagnostics, V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology,
Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia, +7(495)438-13-41
Leyla V. Adamyan, Dr. Med. Sci., Professor, Academician of the Russian Academy of Sciences, Deputy Director for Research, Head of the Gynecological Department,
V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia,
l_adamyan@oparina4.ru, https://orcid.org/0000-0002-3253-4512
Dmitry Yu. Trofimov, Dr. Bio.Sci., Professor, Corresponding Member of the Russian Academy of Sciences, Director of the Institute of Reproductive Genetics,
V.I. Kulakov National Medical Research Center of Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 4 Ac. Oparin str., Moscow, 117997, Russia, +7(495)438-49-51, d_trofimov@oparina4.ru, https://orcid.org/0000-0002-1569-8486
Corresponding author: Maria V. Kuznetsova, mkarja@mail.ru



