Detection of antibodies to SARS-CoV-2 in healthcare professionals of the National Center during the COVID-19 pandemic (April–June 2020)
Objective. To identify IgG antibodies to SARS-CoV-2 in healthcare professionals of the National Medical Research Center for Obstetrics, Gynecology and Perinatology during the quarantine from April to June 2020.Krechetova L.V., Vtorushina V.V., Inviyaeva E.V., Ivanets T.Yu., Donnikov A.Yu., Dolgushina N.V., Sukhikh G.T.
Materials and methods. The study included 1589 healthcare workers: 1293 professionals of ‘green zone’ and 926 medical staff of ‘red zone’. IgG antibodies to SARS-CoV-2 in blood serum were determined using SARS-CoV-2-IgG-ELISA kits (National Hematology Research Center, Russia). SARS-CoV-2 RNA was extracted from nasopharyngeal swabs using the kit PROBA-NK (DNA-technology, LLC, Russia). The virus was identified by RT-PCR using SARS-CoV-2/SARS-CoV Multiplex REAL-TIME PCR Detection Kit (DNA-technology, LLC, Russia).
Results. IgG antibodies to SARS-CoV-2 were detected in 141 healthcare workers (8.9%), controversial results were revealed in 2 professionals, and 1445 (90.9%) workers had no antibodies, including 46 (3.2%) people who had the clinical symptoms of acute respiratory viral disease (ARVI) and identified SARS-CoV-2 RNA. Among healthcare workers with antibodies, the clinical symptoms of ARVI were revealed in 129 (91.5%) workers, they were also detected SARS-CoV-2 RNA; 23 (17.8%) people had clinical symptoms of ARVI but SARS-CoV-2 RNA was not extracted; 12 (8,5%) workers had neither clinical symptoms of ARVI nor detected SARS-CoV-2 RNA.
Conclusion. The presence of IgG antibodies and the absence of SARS-CoV-2 RNA in the nasopharyngeal swab as well as clinical symptoms of the disease may be suggestive of the fact that the worker had this disease; the presence of IgG antibodies and the absence of SARS-CoV-2 RNA as well as clinical symptoms of the disease may be suggestive of the fact that the worker has this disease.
Keywords
In March 2020, the World Health Organization declared a pandemic of a novel viral infection caused by a new strain of β-coronavirus, which may result in severe acute respiratory syndrome similar to the syndrome caused by β-coronavirus strain SARS-CoV-1 (Severe Acute Respiratory Syndrome Related Coronavirus-1) that occurred in 2002–2003. The new strain has been identified as SARS-CoV-2, and the disease caused by this strain is COVID-19 (COronaVIrus Disease 2019). Quarantine measures were introduced due to the lack of acquired immunity to the new pathogen, the susceptibility of the population of all ages to it, the absence of markers of the severity of the disease, as well as the lack of specific therapy; some of the existing hospitals were repurposed and divided into clean («green») and contagious («red») zones.
In April–June 2020, an infectious hospital with 190 beds was established in the National Medical Research Center for Obstetrics, Gynecology and Perinatology, Moscow (hereinafter referred to as the Center) for receiving patients diagnosed with COVID-19.
Healthcare professionals in the red zone of the Center came into contact with patients in special personal protective equipment, which is considered to minimize the possibility of infection; their way of life was supposed to minimize the possibility of external contacts. The medical staff of the green zone wore masks and gloves daily both in the workplace and in public places. This regime was followed for three months until the infectious diseases hospital stopped working. At the end of the quarantine, it was necessary to sum up the results, in particular, to find out whether it was possible to register the signs of the development of specific antiviral immunity in healthcare workers of the Center.
To date, there are no generally accepted criteria for evaluating the effectiveness of developing the memory of innate and adaptive immunity to viral antigens, which can be used in routine clinical practice. The existing stereotypes in understanding the significance of seroconversion and the protective role of antibodies in the elimination of the pathogen and the development of immunological memory lead to the fact that the main focus in assessing immune reactivity for the SARS-CoV-2 virus is to identify antibodies to its most conservative antigens.
The aim of this study was to identify IgG antibodies to SARS-CoV-2 in healthcare professionals of the Center during the quarantine from April to June in 2020.
Materials and Methods
The study included 1589 healthcare workers: 1293 professionals of green zone and 926 medical staff of red zone. The study included the staff of both medical and non-medical specialties on the basis of their desire to be tested for antibodies. Informed consent was obtained from all tested workers.
Blood was collected once from the ulnar vein on an empty stomach.
Detection of IgG antibodies to SARS-CoV-2 in blood serum was carried out by SARS-CoV-2-IgG-ELISA reagent kits produced by National Medical Hematology Research Center, Moscow, Russia. According to the manufacturer’s instructions, the test is intended to perform qualitative and semi-quantitative assessment of antibodies. Positive index (IP) is used to interpret the result, which is calculated using the formula: IP = OD of the sample/Cut-off, where OD of the sample is the value of the optical density of the sample. If IP>1.1, the sample is positive, and if IP <0.9, the sample is negative. If the IP value is between 0.9 and 1.1, the result is doubtful. Samples with doubtful results were re-tested after 1–2 days using a different portion of blood.
The material for detecting SARS-CoV-2 RNA was obtained from the oropharynx using disposable probes. The virus RNA was isolated using a reagent kit PROBANK (LLC, DNA-Technology, Russia).
The virus was identified using a reagent kit for detecting RNA of coronaviruses SARS-CoV-2 and those similar to SARS-CoV by reverse transcription and real-time polymerase chain reaction (RT-PCR) (SARS-CoV-2/SARS-CoV) (LLC, DNA-Technology, Russia). Three genome regions were selected as targets: regions of the N and E gene which are specific to SARS-CoV-2, as well as a conservative region of the E gene that is common to a group of coronaviruses similar to SARS-CoV (including SARS-CoV and SARS-CoV-2). Amplification was performed using amplifier DT-964 (LLC, DNA-Technology, Russia). The results were processed automatically using the software of the amplifier.
Results and Discussion
IgG antibodies to SARS-CoV-2 were detected in 141 healthcare workers (8.9%); doubtful results were revealed in two professionals: one of them showed the clinical symptoms of acute respiratory viral infection (ARVI) and had a positive RT-PCR result, while the other showed no clinical symptoms of ARVI and had a negative RT-PCR result.
IgG antibodies to SARS-CoV-2 were not detected in 1445 (90.9%) workers. Among them 46 (3.2%) people had the clinical symptoms of (ARVI) and identified SARS-CoV-2 RNA in oropharyngeal swabs.
Among healthcare workers with antibodies, the clinical symptoms of ARVI were revealed in 129 (91.5%) workers, they were also detected SARS-CoV-2 RNA; 23 (17.8%) people had clinical symptoms of ARVI but SARS-CoV-2 RNA was not extracted; 12 (8,5%) workers had neither clinical symptoms of ARVI nor detected SARS-CoV-2 RNA (including 3 healthcare workers who were in contact with a COVID-19 patient). When comparing the content of antibodies in the blood of medical staff of the red and green zones, the following facts were noted: 130 (10.1%) people out of 1293 tested workers of the green zone had COVID-19, including 127 people who had antibodies, and 3 workers with doubtful results; 14 (5.2%) people out of 269 tested medical staff of the red zone had COVID-19, which is half as much as in the green zone, antibodies were detected in all those who were ill.
According to the published data on the results of antibody research in patients with COVID-19 in Wuhan (China), seroconversion in SARS-CoV-2 infection is characterized by almost simultaneous registration of IgM and IgG antibodies in the blood 3–5 days after the onset of SARS symptoms, the dynamics of the increase of both classes of antibodies is the same for almost three weeks [1], after which the level of IgM antibodies decreases and only IgG antibodies are registered.
Sethuraman N. et al. [2] in their study published in June 2020 analyzed the dynamics of registration of the SARS-CoV-2 virus in PCR and registration of IgM and IgG antibodies as related to the manifestation of SARS symptoms. They showed that virus RNA can be detected a week before the onset of SARS symptoms, and it can be detected for a month after the manifestation of the disease. The absence of virus RNA in the biomaterial coincides with a decrease in the level of IgM antibodies. It is noted that the level of antibodies does not correlate with the severity of the disease. The lack of IgM antibody production in the patients with COVID-19 is mentioned in some publications [3], but this result may be associated with the low sensitivity of the test systems. Nowadays, all the existing ELISA kits of different manufacturers for detecting IgM and IgG antibodies are semi-quantitative, quantitative tests cannot be used due to the lack of standards for antibodies. It is also impossible to carry out a routine assessment of the protective function performed by antibodies, i.e. virus neutralizing capacity cannot be evaluated, as today it can be assessed only in vitro in cell culture infected with a virus. The level of antibodies with virus neutralizing capacity has been shown to be very low in some convalescents [4].
Moreover, the difficulties and peculiarities of interpreting the reactivity of humoral antiviral immunity are associated with the plasticity of virus genomes, i.e. with high variability resulting from spontaneous mutations or homologous recombination with genomes of related species. The genome structure of coronaviruses circulating in natural reservoirs is always mosaic, and the coronaviruses themselves exist as quasi-individual pools. As a rule, infection is caused by a quasi-individual which is a set of genomes of homologous species [5].
However, the phenomenon of antibody-dependent enhancement (ADE) of viral infection was observed in model experiments. This phenomenon has been known since 1964. To date, it has been noted in a number of animal and human virus families, including RNA-containing viruses such as Retroviridae, Picornaviridae (causative agents of polio, rhinitis, hepatitis), Orthomyxoviridae (causative agents of influenza A), Ebolavirus, and DNA-containing viruses, namely Parvoviridae, Epstein-Barr virus, Herpesviridae [6, 7]. This phenomenon may also develop for SARS-CoV-2 [8, 9]. The proposed mechanisms of ADE are related to the interaction of antigen-antibody complexes via Fc fragments of immunoglobulins with their receptors on the cell surface and proteins of the complement system (Fig. 1).
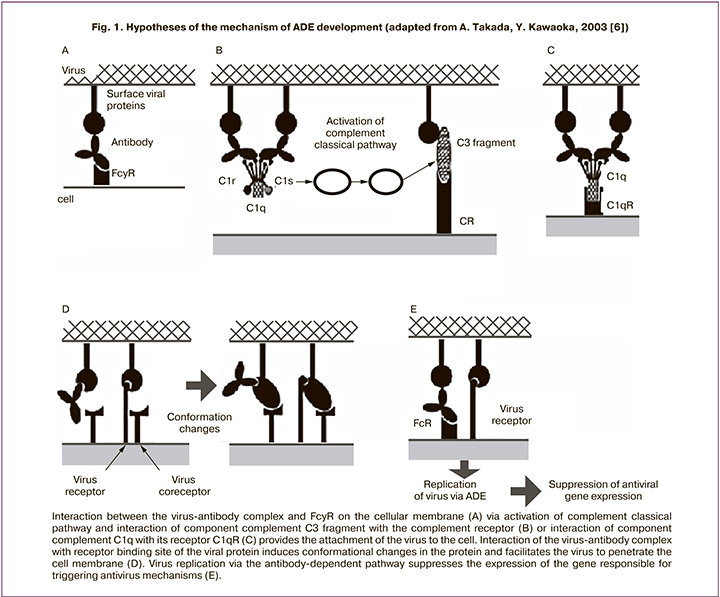
The Fc fragment is used in the physiological effects of immunoglobulins such as opsonization (pathogen labeling), lysis of pathogen-loaded cells, and degranulation of mast cells, basophils, and eosinophils. Receptors to the Fc fragment of immunoglobulin or to the specified complement components are present on all immunocompetent cells, including T- and B-lymphocytes, dendritic cells, natural killer cells, macrophages, neutrophils, eosinophils, basophils, as well as human platelets, mast cells. The mechanisms of ADE are supposed to involve complexes with a low-affinity antigen-antibody bond (these can be both multivalent IgM antibodies and natural antibodies). The virus enters the cell through the Fc fragment of immunoglobulin, but the breakdown of such low-affinity complexes in cells does not lead to the activation of antiviral metabolic mechanisms. On the contrary, viral particles released from such complexes invade the metabolic pathways of the cell, replicate, and reactivate. When resulting in the death of the host cell, they release various biologically active compounds into the intercellular space, which ultimately cause the development of pathological reactions leading to a complex of symptoms of a systemic inflammatory response characteristic of patients with severe COVID-19 (including «cytokine storm»). Currently, quantitative assessment of such complexes is not possible in routine practice.
The participation of antibodies in the pathogenesis of increased severity of COVID-19 is discussed in the study [10], which reviews immunological changes in viral pneumonia caused by coronaviruses strains SARS-CoV-1, MERS-CoV, SARS-CoV-2, and summarizes the results of treating patients with severe respiratory failure using combined therapy with highdose immunoglobulins administered intravenously in combination with low-molecular-weight heparins (Fig. 2).
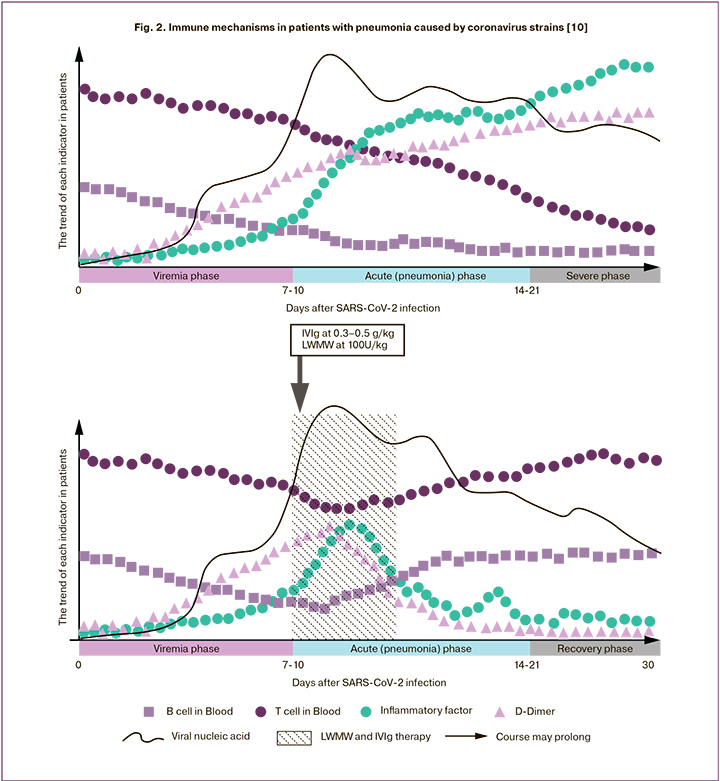
Based on the above, there is a difficulty in interpreting the results of the studies of IgG antibodies in the blood of the medical staff of the Center, first of all, due to the lack of information about a specific strain of the pathogen, the lack of quantitative evaluation of antibodies and their virus-neutralizing ability. It is important to assess the duration of antibodies presence in the blood, the possibility of re-infection if they are detected in the blood, and the severity of re-infection. The use of convalescent plasma for the treatment of severe forms of coronavirus infection is debatable, since the criteria for specific donation have not been developed, although examples of the use of convalescent plasma have already been published [11–13]. It is essential to evaluate immune reactivity in the patients whose recovery was not accompanied by the production of IgG antibodies to SARS-CoV-2. In addition, in the course of our study, there were twice as many cases in the red zone (5.2%), compared to the green one (10.1%), which may indicate the effectiveness of the personal protective equipment and the ways it was used. The answer to this question also requires further research.
Conclusion
Thus, based on the study of antibodies in the healthcare professionals of the Center and the literature data, the following conclusion can be made: the presence of IgG antibodies and the absence of SARS-CoV-2 RNA in the nasopharyngeal swab as well as clinical symptoms of the disease are suggestive of the fact that the worker had this disease; the presence of IgG antibodies and the absence of SARS-CoV-2 RNA and the presence of clinical symptoms of the disease may be suggestive of the fact that the worker has this disease, but not about its stage. IgG antibodies in asymptomatic patients and in patients with a mild form of the disease may not be revealed during the entire period of detecting the pathogen RNA in the nasopharyngeal swab and after recovery. The presence of IgG antibodies and the absence of the pathogen RNA in the swab a month after the onset of the disease is not an indicator that the patient poses risk to others and, therefore, is not a reason for self-isolation and restriction of contacts.
References
- Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J. et al. Antibody detection and dynamic characteristics in patients with COVID-19. Clin. Infect. Dis. 2020; Apr. 19: ciaa461. https://dx.doi.org/10.1093/cid/ciaa461.
- Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020; 323(22): 2249-51. https://dx.doi.org/10.1001/jama.2020.8259.
- Padoan A., Sciacovelli L., Basso D., Negrini D., Zuin S., Cosma C. et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: A longitudinal study. Clin. Chim. Acta. 2020; 507: 164-6. https://dx.doi.org/10.1016/j.cca.2020.04.026.
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S. et al. Neutralizing antibody responses to SARS-Cov-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020: 2020.03.30.20047365.
- Супотницкий М.В. Новый коронавирус SARS-CoV-2 в аспекте глобальной эпидемиологии коронавирусных инфекций. Вестник войск РХБ защиты. 2020; 4(1): 32-65. [M.V. Supotnitskiy. Novel coronavirus SARS-CoV-2 in the context of global epidemiology of coronavirus infections. Journal of NBC Protection Corps. 2020; 4(1): 32-65. (in Russian)]. https://dx.doi.org/10.35825/2587-5728-2020-4-1-32-65.
- Takada A., Kawaoka Y. Antibody-dependent enhancement of viral infection: molecular mechanisms and in vivo implications. Rev. Med. Virol. 2003; 13(6): 387-98. https://dx.doi.org/10.1002/rmv.405.
- Wang S.F., Tseng S.P., Yen C.H.,Yang J.Y., Tsao C.H., Shen C.W. et al. Antibody-dependent SARS coronavirus infection is mewdiated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014; 451(2): 208-14. https://dx.doi.org/10.1016/j.bbrc.2014.07.090.
- Ulrich H., Pillat M.M., Tárnok A. Dengue fever, COVID‐19 (SARS‐CoV‐2), and antibody‐dependent enhancement (ADE): a perspective. Cytometry A. 2020; Jun. 7. https://dx.doi.org/10.1002/cyto.a.24047.
- Peron J.P.S., Nakaya H. Susceptibility of the elderly to SARS-CoV-2 infection: ACE-2 overexpression, shedding, and antibody-dependent enhancement (ADE). Clinics (Sao Paulo). 2020; 75: e1912. https://dx.doi.org/10.6061/clinics/2020/e1912.
- Ling L., Lianfeng L., Wei C., Taisheng L. Hypothesis for potential pathogenesis of SARS-CoV-2 infection–a review of immune changes in patients with viral pneumonia. Emerg. Microb. Infect. 2020; 9(1): 727-32. https://dx.doi.org/10.1080/22221751.2020.1746199.
- Ye M., Fu D., Ren Y., Wang F., Wang D., Zhang F. et al. Treatment with convalescent plasma for COVID-19 patients in Wuhan. J. Med. Virol. 2020; Apr. 15. https://dx.doi.org/10.1002/jmv.25882.
- Tiberghien P., de Lamballerie X., Morel P., Gallian P., Lacombe K., Yazdanpanah Y. Collecting and evaluating convalescent plasma for COVID-19 treatment: Why and How? Review. Vox Sang. 2020; Apr. 2. https://dx.doi.org/10.1111/vox.12926.
- Barone P., DeSimone R.A. Convalescent plasma to treat coronavirus disease 2019 (COVID-19): considerations for clinical trial design. Review. Transfusion. 2020; 60(6): 1123-7. https://dx.doi.org/10.1111/trf.15843.
Received 07.07.2020
Accepted 10.07.2020
About the Authors
Lubov V. Krechetova, Doctor of Science, MD, PhD, Head of the Laboratory of Clinical Immunology, National Medical Research Center for Obstetrics, Gynecology and Perinatology Ministry of Healthcare of the Russian Federation. Tel.: +7(495)438-11-83. E-mail: l_krechetova@oparina4.ru.117997, Russia, Moscow, Academician Oparin str., 4.
Valentina V. Vtorushina, MD, PhD, immunologist-allergist doctor, National Medical Research Center for Obstetrics, Gynecology and Perinatology Ministry
of Healthcare of the Russian Federation. Tel.: +7(916)980-78-95. E-mail: vtorushina@inbox.ru.
117997, Russia, Moscow, Academician Oparin str., 4.
Eugenia V. Inviyaeva, PhD, Senior Researcher of the Laboratory of Clinical Immunology, National Medical Research Center for Obstetrics, Gynecology and Perinatology Ministry of Healthcare of the Russian Federation. Tel.: +7(495)438-11-83. E-mail e_inviyaeva@oparina4.ru.
117997, Russia, Moscow, Academician Oparin str., 4.
Tatiana Yu. Ivanets, Doctor of Science, MD, PhD, Head of the Clinical Diagnostic Laboratory, National Medical Research Center for Obstetrics, Gynecology and Perinatology Ministry of Healthcare of the Russian Federation. Tel.: +7(910)404-26-69. Е-mail: t_ivanets@oparina4.ru.
117997, Russia, Moscow, Academician Oparin str., 4.
Andrey E. Donnikov, MD, PhD, Head of Laboratory of molecular genetic methods, National Medical Research Center for Obstetrics, Gynecology and Perinatology Ministry
of Healthcare of the Russian Federation. Tel.: +7(495)438-49-51. E-mail: a_donnikov@oparina4.ru.
117997, Russia, Moscow, Academician Oparin str., 4.
Nataliya V. Dolgushina, Doctor of Science, MD., PhD., M.P.H., Head of R&D Department, National Medical Research Center for Obstetrics, Gynecology and Perinatology Ministry of Healthcare of the Russian Federation. Tel.: +7(495)438-49-77. E-mail: n_dolgushina@oparina4.ru.
117997, Russia, Moscow, Academician Oparin str., 4.
Gennady T. Sukhikh, MD, PhD, Professor, Academician of Russian Academy of Sciences, Director of National Medical Research Center for Obstetrics, Gynecology and Perinatology Ministry of Healthcare of the Russian Federation. Tel.: +7(495)438-18-00. E-mail: gtsukhih@mail.ru.
117997, Russia, Moscow, Academician Oparin str., 4.
For citing: Krechetova L.V., Vtorushina V.V., Inviyaeva E.V., Ivanets T.Yu.: Donnikov A.Yu., Dolgushina N.V., Sukhikh G.T. Detection of antibodies to SARS-CoV-2 in healthcare professionals of the National Center during the COVID-19 pandemic (April–June 2020).
Akusherstvo i Ginekologiya / Obstetrics and Gynecology. 2020; 7: 122-128 (in Russian)
https://dx.doi.org/10.18565/aig.2020.7.122-128



