Vaginal microbiome in pregnant women with preterm prelabor rupture of membranes at 22–28 weeks' gestation
Aim. To investigate vaginal microbiome in pregnant women with preterm prelabor rupture of membranes (PPROM) before 28 weeks' gestation using metagenomic sequencing.Kuznetsova N.B., Bushtyreva I.O., Dybova V.S., Barinova V.V., Polev D.E., Aseev M.V., Dudurich V.V.
Materials and methods. The study comprised 55 pregnant women, including patients with PPRPO before 28 weeks' gestation (group I) and women with a healthy pregnancy (group II).
Results. Statistically significant between-group differences were found in the relative abundance of microorganisms for the vaginal microbiome regarding 13 genera of microorganisms including Lactobacillus, Pediococcus, Prevotella, Prevotella 6, Peptostreptococcus, Peptoniphilus, Anaerococcus, Fusobacterium, Parvimonas, Lawsonella, Sutterella, Mobiluncus, and Thermus. Also, there were statistically significant differences in detection rates of microorganisms' genera, including Pediococcus, Prevotella, Prevotella 6, Anaerococcus, Peptoniphilus, Parvimonas, Lawsonella, Sutterella, Mobiluncus, Thermus.
Conclusion. The presence of bacteria of the genus Prevotella (especially Prevotella 6), Anaerococcus, Peptoniphilus, Parvimonas, Lawsonella, Sutterella, Mobiluncus, Thermus in the vaginal microbiome of pregnant women with PPRPO, and low relative abundance of bacteria of the genus Lactobacillus and Pediococcus during pregnancy are associated with preterm prelabor rupture of membranes before 28 weeks' gestation, which warrants further investigations.
Keywords
The international Human Microbiome Project allowed for a fresh look at the role of microorganisms in human physiology and pathology [1], resulting in the concept of the microbiome. Sequencing technologies have helped to expand our knowledge of the vaginal microbiota, obtained to this day mainly through culture and DNA amplification.
The research on the role of the vaginal microbiome in pregnancy complications, including preterm prelabor rupture of membranes (PPROM), has marked clinical importance. This is extremely important because standard detection procedures such as culture and polymerase chain reaction have been exhausted. A fundamentally new approach could be the characterization of the vaginal microbiota using 16S rRNA gene sequence analysis. Since the vagina is one of the most accessible organs for studying the human body's biotopes, today, there is a sufficient amount of data on the vaginal microbiome in women of different races under normal conditions and pathological settings [2–5]. However, investigating the role of the vaginal microbiome during pregnancy leading to PPROM is a challenging problem.
Given the potentially significant role of the infectious-inflammatory factor in the development of PPROM and the problem of antibiotic resistance, expanding knowledge of the microbiome using 16s rRNA sequencing techniques is critical for developing strategies aimed at preventing adverse outcomes in PPROM.
This study aimed to investigate vaginal microbiome in pregnant women with PPROM before 28 weeks' gestation using metagenomic sequencing.
Materials and methods
The study was conducted at the Rostov Regional Perinatal Center between 01.06.2019 and 31.03.2020. It comprised 55 pregnant women, including patients with PPRPO before 28 weeks' gestation (group I) and women with a healthy pregnancy (group II).
Inclusion criteria were age from 18 to 40 years and Caucasian race. Exclusion criteria were pregnancy resulting from assisted reproductive technologies, multiple-gestation pregnancy, polyhydramnios, surgery on the cervix, congenital malformations of the uterus, fetal malformations, human immunodeficiency virus infection, hepatitis B, hepatitis C, syphilis, severe non-obstetric pathology, gestational diabetes, antibacterial therapy during pregnancy before inclusion in the study, invasive procedures (amniocentesis, amnioreduction), trauma during pregnancy, unhealthy behaviors (smoking, alcohol abuse).
All participants provided informed consent to participate in the study. The Research Ethics Committee of Rostov State Medical University approved this study.
The PPROM diagnosis was made under the existing diagnostic criteria, including the visualization of amniotic fluid passing from the cervical canal during speculum examination and cough test, ultrasonic measurement of the amniotic fluid index, positive amniotest, and positive AmniSure test.
In group I, a sample of vaginal fluid was collected under direct visualization from the posterior vaginal fornix during the first 12 hours after the onset of amniotic fluid discharge and before starting antibiotic therapy at 22 to 28 weeks' gestation. In group II, samples of vaginal fluid were collected at similar gestational age (22 weeks –27 weeks six days).
The samples were placed in sterile tubes with a transport medium "transport medium with mucolytic" (Central Research Institute of Epidemiology, Russia) and stored at + 4℃ until DNA isolation.
Total DNA was isolated from the samples using the RIBO-prep kit (Central Research Institute of Epidemiology, Russia) per the manufacturer's instructions.
16S DNA libraries were prepared according to the Illumina 16S Metagenomic Sequencing Library Preparation protocol (Part # 15044223 Rev.B). Bioinformatic analysis of sequencing data was carried out using programming languages R v.3.6 [6] and Python 3.6.9. At the first stage, the primer sequences were truncated from the beginning of the paired final reads, and the pairs of reads without the primer sequence were discarded. The final 25 base pairs were then truncated from the end of each read as low-quality bases and the resulting data processed using the DADA2 workflow to identify sequence variants [7] accurately. After the derivation of the exact sequence variants, forward and backward readings were combined by concatenation. The resulting sequences were used for comparison according to Bayesian taxonomic classification using the SILVA v132 database as a reference [8]. The species assignment was performed using the exact match algorithm in DADA2 using SILVA v132 sequences preprocessed accordingly using custom scripts.
Statistical analysis
Between-group differences in quantitative variables showing normal distribution (age of patients, body mass index, gestational age at the time of biological material collection) were assessed using the Student's t-test for independent samples. Numerical variables that were not normally distributed (comparison of the relative abundance of bacteria (of the total composition)) were compared by the Mann– Whitney U-test. The distribution of continuous variables was tested for normality using the Kolmogorov–Smirnov test. Categorical variables (frequencies and percentages of anamnestic characteristics and occurrence of bacteria) were compared using Fisher's exact test. To identify a possible relationship between the indicators, a correlation analysis was carried out to determine the Spearman correlation coefficients.
The differences were considered statistically significant at p<0.05. Calculations were performed in R (version 3.6, R Foundation for Statistical Computing, Vienna, Austria).
Results
Groups I and II included 27 and 28 women, respectively, who met the study inclusion criteria. The mean gestational age of women in the group I and II at the time of amniotic fluid discharge was 25.6 ± 1.4 25.8 ± 2.2 weeks, respectively. The mean gestational age of women in the group I and II at the time of inclusion in the study was 25.8 ± 2.2 weeks. The mean gestational age of women in the group I and II was 29 ± 5.3 and 29 ± 4.1 years.
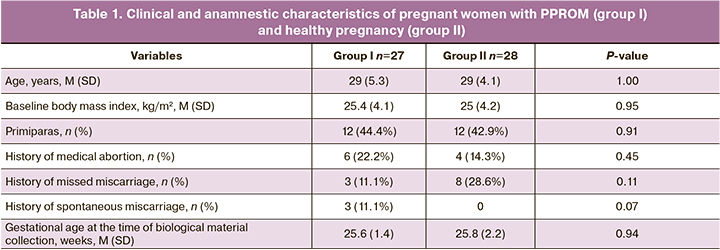
There was no statistically significant difference in obstetric and gynecological history (medical abortions, missed miscarriages, and spontaneous abortions) when comparing the two groups (Table 1).
All patients of group I were examined following the Order of the Ministry of Health of Russia dated 01.11.2012 No. 572 n “On approval of the Procedure for the provision of medical care in the profile Obstetrics and Gynecology (except for the use of assisted reproductive technologies).” The management of pregnant women with PPROM at admission was provided under the protocol of the Ministry of Health of Russia dated December 17, 2013, N. 15-4/10/2-9480.
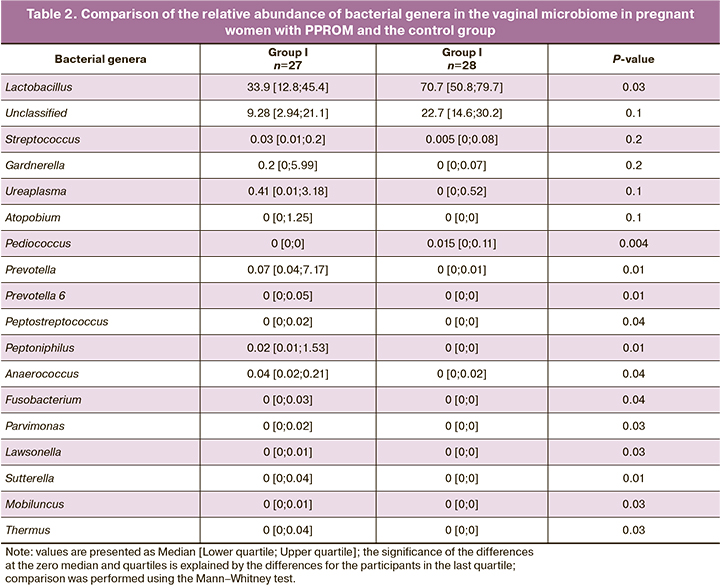
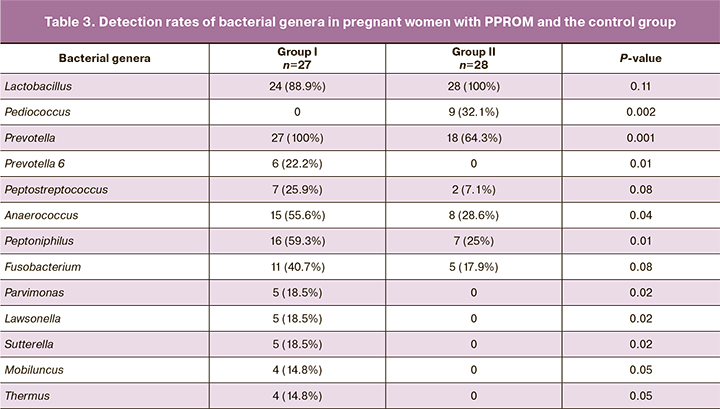
Vaginal microbiota was analyzed using gene sequencing technology, including massively parallel sequencing (new generation sequencing) of the 16S rRNA gene and bioinformatics analysis of the data obtained with DNA classification by taxonomic units. The vaginal microbiome was analyzed by identification of microorganisms to the genus and species level. The analysis was conducted by comparing relative abundance (Table 2) and detection rates (Table 3) of microorganisms in both groups.
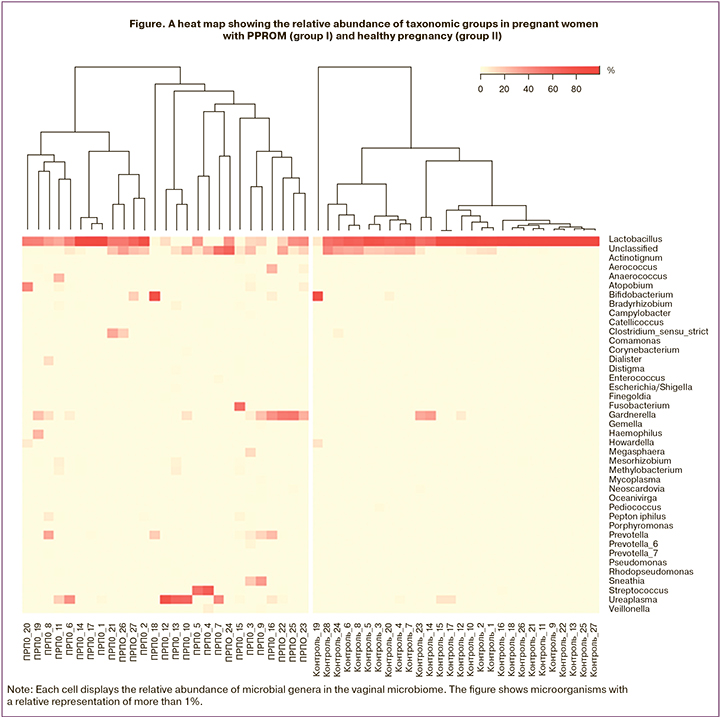
Analysis of relative abundance showed that the vaginal microbiome was dominated by bacteria from the genus Lactobacillus (Figure).
Statistically significant differences in relative abundance were found for 13 genera of microorganisms in the vaginal microbiome (Table 2).
The relative representation of microorganisms of the genus Lactobacillus in the vaginal microbiome in pregnant women with PPROM before 28 weeks (group I) and healthy pregnancy (group II) was 33.9 (12.8; 45.4) and 70.7 (50.8; 79.7), respectively, p=0.03. Also, in pregnant women with a healthy pregnancy (group II), the relative abundance of microorganisms of the genus Pediococcus was higher than 0.015 [0; 0.11], compared with pregnant women with PPROM (group I) – 0 [0; 0], p=0.004.
The microorganisms with higher relative abundance in pregnant women with PPROM before 28 weeks (group I) included anaerobic bacteria of the genera Prevotella, Prevotella 6, Peptostreptococcus, Peptoniphilus, Anaerococcus, Fusobacterium, Parvimonas, Lawsonella, Sutterella, and Mobiluncus, Table ). Of note is the very low relative representation of all genera of microorganisms in the vaginal microbiome except those belonging to the genus Lactobacillus.
Comparison of microorganism detection rates (Table 3) in the vaginal microbiome also showed a statistically significantly lower occurrence of bacteria of the genus Pediococcus in pregnant women with PPROM than in women in group II (healthy pregnancy), p = 0.002.
Only detection rates of bacteria of the genera Prevotella, Anaerococcus, Peptoniphilus, Parvimonas, Lawsonella, Sutterella, Mobiluncus, and Thermus were found to be higher in pregnant women with PPROM (Table 3). In particular, microorganisms of the genus Prevotella were detected in all samples of women with PPROM (100%). In the control group, bacteria of the genus Prevotella were detected in 64.1% (18) of women, p=0.001. Four additional groups of genus-level sequences were also identified and assigned unique designations, including Prevotella 2, Prevotella 6, Prevotella 7, and Prevotella 9. Among these, statistically significant differences were noted for Prevotella 6, which was found in 6 women with PPROM but not found in group II, p=0.01. Also, in group I, bacteria of the genera Peptoniphilius (p=0.01) and Anaerococcus (p=0.04) were quite common. The rest of the microorganisms (bacteria of the genera Parvimonas, Lawsonella, Sutterella), which had significantly different detection rates in the study groups, were observed less often and were found only in group I (Table 3).
The duration of ruptured membranes varied from 49 to 162 hours and averaged 97 hours (4 days). There was no correlation between the duration of ruptured membranes and vaginal microbiome composition. The indications for delivery in group I were the onset of spontaneous preterm labor [66.7% (n=18)], anhydramnios [22.2% (n=6)], and chorioamnionitis [11.1% (n=3)]. Eighteen (66.6%) and 9 (33.3%) women had vaginal and cesarean deliveries, respectively.
Discussion
Although the vaginal microbiome is one of the most-studied topics, due to the availability of this biotope of the human body for sampling, the characteristics of the vaginal microbiome in pregnant women with PPROM and during the latency period before the onset of labor is a practically unexplored area of knowledge.
Our findings suggest a low abundance of Lactobacilli in the microbiome of women with PPROM, compared with patients with a healthy pregnancy. Reduced Lactobacilli abundance creates an acidic environment (pH <4.5), and along with many other factors plays a crucial role in destabilizing the vaginal microbiome. On the one hand, these changes may be associated with the effect of continually leaking amniotic fluid, which, due to its alkaline environment, physically wash out and chemically suppress the growth of Lactobacilli [2, 4]. The low abundance of Lactobacilli results in colonization of an accessible niche by opportunistic pathogens, which leads to dysbiosis against the background of PPROM. On the other hand, this condition may be a consequence of bacterial vaginosis preceding rupture of the membranes. Either way, the processes that occur after the onset of amniotic fluid leakage have potentially adverse consequences.
It is essential to mention that lactobacilli's simple dominance in the vaginal microbiome does not guarantee a favorable pregnancy outcome. According to our study, in pregnant women with PPROM, vaginal flora was dominated by L. iners. In group II, L. crispatus, L. acidophilus, L. jensenii, L. gasseri were more often observed, which is similar to the data reported by Z.S. Khodzhaeva et al. [9]. L. iners is a dominant part of the vaginal microbiota in the transitional phase between normal and abnormal that may predispose to abnormal vaginal microflora [2, 10]. Some studies have also suggested that L. iners in a dominant state might be associated with preterm delivery [3]. In contrast to L.iners, the dominance of L. crispatus is more often observed in women who had a term delivery [4].
Along with the difference in the abundance of lactobacilli, bacteria of the genus Pediococcus were not detected in pregnant women with PPROM, in contrast to pregnant women in group II. These bacteria belong to the genus of gram-positive lactic acid bacteria belonging to the Lactobacillaceae family. Due to pediocin and bacteriocin release, they have antimicrobial and probiotic properties [11, 12], thus contributing to successful pregnancy completion.
Of particular interest is the predominance of anaerobic bacteria in the microbiome of women with PPROM, which have been described as opportunistic microorganisms capable of causing vaginal dysbiosis [13]. In particular, the most common microorganisms in group I was bacteria of the genus Prevotella. Four other genus-level groups belonging to Prevotella were identified, including Prevotella 2, Prevotella 6, Prevotella 7, and Prevotella 9, similar to earlier studies [14], which made it possible to reveal differences in the abundance of Prevotella 6. The available literature also indicates a potential role of Prevotella in the etiology of PPROM. Prevotella is known to be associated with bacterial vaginosis and preterm labor [5]. Considering the supposed role of this genus in synergistic relationships with microorganisms associated with bacterial vaginosis and their production of lipopolysaccharides in the vaginal environment, the prevalence and diversity of Prevotella bacteria in the microbiome of pregnant women with PPROM undoubtedly requires an in-depth study.
Like Prevotella, bacteria of the genus Peptostreptococcus are considered clinically significant anaerobic cocci. The only representative of this genus in our study was the species Peptostreptococcus anaerobius. According to the available data, Peptostreptococcus anaerobius is found in both vaginal and placental samples of pregnant women, associated with more unfavorable outcomes and lower birth weight and height of newborns [15]. It should be noted that some species of bacteria of the genus Peptostreptococcus are now divided into a separate group and belong to the genus Peptoniphilius and the genus Anaerococcus. However, associations with an unfavorable pregnancy course, similar to bacteria of the genus Peptostreptococcus, persist [14], which is consistent with the results of our study.
It will also be of interest to study bacteria from anatomical sites atypical for the reproductive system (bacteria of the genera Fusobacterium, Parvimonas, Lawsonella). The primary human body habitat of bacteria of the genus Fusobacterium is the gastrointestinal tract, including the oral cavity and periodontium. Due to the study of the human microbiome, F. nucleatum was found in tissues of the placenta and fetus, including amniotic fluid [16], umbilical cord blood [17], and in gastric aspirates of newborns [18] in women whose pregnancy was complicated by preterm labor without PPROM. The strains of F. nucleatum identified in the amniotic fluid and placenta correspond to strains from the oral cavity, not the lower genital tract, which confirms the involvement of oral facultative species such as F. nucleatum in adverse pregnancy outcomes due to the hematogenous spread of microorganisms [19].
Bacteria of the genera Parvimonas and Lawsonella are other representatives of the atypical anatomical sites found in the reproductive tract. The only representative of bacteria of the genus Parvimonas in our study was the species Parvimonas micra, and the bacteria of the genus Lawsonella was the species Lawsonella clevelandensis. As one of the inhabitants of dental plaque, the species Parvimonas micra in patients with chronic periodontitis was found in bacterial vaginosis [20] and chorioamnionitis [21]. The species Lawsonella clevelandensis, which, according to the literature, is also identified as part of the oral microbiota, is rarely associated with human infections [22]. However, the literature reported 8 cases of complicated infection with this microorganism [23].
In our study, anaerobic microorganisms were represented by bacteria of the genus Sutterella found in the group I patients. Current literature provides no evidence on the relationship of bacteria of the genus Sutterella with the unfavorable course and pregnancy outcome [24]. However, there is an increase in their concentration in the vagina and cervical canal with sexually transmitted infections [25]. The role of this microorganism in adverse pregnancy outcomes remains to be established in further studies.
It is also worth commenting on detecting bacteria of the genera Mobiluncus and Thermus in group I. Each of the microorganisms was represented by Mobiluncus curtisii and Thermus scotoductus, respectively. The exact role of these bacteria in the pathogenesis of dysbiosis and other pregnancy complications has not been established. The dominance of Mobiluncus in the vagina is observed in pregnant women with asymptomatic bacterial vaginosis [26] and in pregnant women with preterm labor [27]. Bacteria of the genus Thermus have been observed as one of the dominant microorganisms in the decidual microbiome during healthy pregnancy [28]. Still, the relationship with possible complications has not been investigated.
Conclusion
A deficiency in lactobacilli and the predominance of anaerobes and higher diversity in the vaginal microbiome in patients with PPROM than in healthy women emphasize pronounced dysbiosis with high individual variability in patients with adverse perinatal outcomes. Further studies using sequencing are needed to investigate the features of the vaginal microbiome before the onset of PPROM since shifts in the vaginal microbial communities can be regarded as one of the immediate causes of preterm labor.
References
- Proctor L., Lo Tempio J., Marquitz A., Daschner P., Xi D., Flores R. et al. NIH Human Microbiome Portfolio Analysis Team. A review of 10 years of human microbiome research activities at the US National Institutes of Health, Fiscal Years 2007-2016. Microbiome. 2019; 7(1): 31. https://dx.doi.org/10.1186/s40168-019-0620-y.
- Paramel Jayaprakash T., Wagner E.C., van Schalkwyk J., Albert A.Y., Hill J.E., Money D.M.; PPROM Study Group. High diversity and variability in the vaginal microbiome in women following preterm premature rupture of membranes (PPROM): a prospective cohort study. PLoS One. 2016; 11(11): e0166794. https://dx.doi.org/10.1371/journal.pone.0166794.
- Petricevic L., Domig K.J., Nierscher F.J., Sandhofer M.J., Fidesser M., Kron-dorfer I. et al. Characterisation of the vaginal Lactobacillus microbiota associated with preterm delivery. Sci. Rep. 2014; 4: 5136. https://dx.doi.org/10.1038/srep05136.
- Romero R., Hassan S.S., Gajer P., Tarca A.L., Fadrosh D.W., Bieda J. et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014; 2: 18. https://dx.doi.org/10.1186/2049-2618-2-18.
- DiGiulio D.B., Callahan B.J., McMurdie P.J., Costello E.K., Lyell D.J., Robaczewska A. et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA. 2015; 112(35): 11060-5. https://dx.doi.org/10.1073/pnas.1502875112.
- R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria; 2018. Available at: https://www.gbif.org/ru/tool/81287/r-a-language-and-environment-for-statistical-computing
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016; 13(7): 581-3. https://dx.doi.org/10.1038/nmeth.3869.
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013; 41(Database issue): D590-6. https://dx.doi.org/10.1093/nar/gks1219.
- Ходжаева З.С., Припутневич Т.В., Муравьева В.В., Гусейнова Г.Э., Горина К.А., Мишина Н.Д. Оценка состава и стабильности микробиоты влагалища у беременных в процессе динамического наблюдения. Акушерство и гинекология. 2019; 7: 30-8. [Khodzhaeva Z.S., Priputnevich T.V., Murav'eva V.V., Guseinova G.E., Gor-ina K.A., Mishina N.D. The composition and stability of the vaginal microbiota in pregnant women during dynamic observation. Obstetrics and ynecology. 2019; 7: 30-8. (in Russian)]. https://dx.doi.org/10.18565/aig.2019.7.30-38.
- Ragaliauskas T., Plečkaitytė M., Jankunec M., Labanauskas L., Baranauskiene L., Valincius G. Inerolysin and vaginolysin, the cytolysins implicated in vaginal dysbiosis, differently impair molecular integrity of phospholipid membranes. Sci. Rep. 2019; 9(1): 10606. https://dx.doi.org/10.1038/s41598-019-47043-5.
- Mehta R., Arya R., Goyal K., Singh M., Sharma A.K. Bio-preservative and therapeutic potential of pediocin: recent trends and future perspectives. Recent Pat. Biotechnol. 2013; 7(3): 172-8. https://dx.doi.org/10.2174/18722083113076660008.
- Kaur B., Garg N., Sachdev A., Kumar B. Effect of the oral intake of probi-otic Pediococcus acidilactici BA28 on Helicobacter pylori causing peptic ulcer in C57BL/6 mice models. Appl. Bochem. Biotechnol. 2014; 172(2): 973-83. https://dx.doi.org/10.1007/s12010-013-0585-4.
- Ходжаева З.С., Гусейнова Г.Э., Муравьева В.В., Донников А.Е., Мишина Н.Д., Припутневич Т.В. Характеристика микробиоты влагалища у беременных с досрочным преждевременным разрывом плодных оболочек. Акушерство и гинекология. 2019; 12: 66-74. [Khodzhaeva Z.S., Guseinova G.E., Muravyeva V.V., Donnikov A.E., Mishina N.D., Priputnevich T.V. Characteristics of the vaginal microbiota in pregnant women with preterm premature rupture of the membranes. Obstetrics and gynecology. 2019; 12: 66-74. (in Russian]. https://dx.doi.org/10.18565/aig.2019.12.66-74.
- Henderson G., Yilmaz P., Kumar S., Forster R.J., Kelly W.J, Leahy S.C. et al. Improved taxonomic assignment of rumen bacterial 16S rRNA sequences using a revised SILVA taxonomic framework. PeerJ. 2019; 7: e6496. https://dx.doi.org/ 10.7717/peerj.6496.
- Doyle R.M., Harris K., Kamiza S., Harjunmaa U., Ashorn U., Nkhoma M. et al. Bacterial communities found in placental tissues are associated with severe chorioamnionitis and adverse birth outcomes. PLoS One. 2017; 12(7): e0180167. https://dx.doi.org/10.1371/journal.pone.0180167.
- Doyle R.M., Alber D.G., Jones H.E., Harris K., Fitzgerald F., Peebles D. et al. Term and preterm labour are associated with distinct microbial community structures in placental membranes which are independent of mode of delivery. Placenta. 2014; 35(12): 1099-101. https://dx.doi.org/10.1016/j.placenta.2014.10.007.
- Wang X., Buhimschi C.S., Temoin S., Bhandari V., Han Y.W., Buhimschi I.A. Comparative microbial analysis of paired amniotic fluid and cord blood from pregnancies complicated by preterm birth and early-onset neonatal sep-sis. PLoS One. 2013; 8(2): e56131. https://dx.doi.org/10.1371/journal.pone.0056131.
- Gonzales-Marin C., Spratt D.A., Allaker R.P. Maternal oral origin of Fuso-bacterium nucleatum in adverse pregnancy outcomes as determined using the 16S-23S rRNA gene intergenic transcribed spacer region. J. Med. Microbiol. 2013; 62(Pt. 1): 133-44. https://dx.doi.org/10.1099/jmm.0.049452-0.
- Vander Haar E.L., So J., Gyamfi-Bannerman C., Han Y.W. Fusobacterium nucleatum and adverse pregnancy outcomes: epidemiological and mechanistic evidence. Anaerobe. 2018; 50: 55-9. https://dx.doi.org/10.1016/j.anaerobe.2018.01.008.
- Romero R., Hassan S.S., Gajer P., Tarca A.L, Fadrosh D.W., Nikita L. et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014; 2(1): 4. https://dx.doi.org/10.1186/2049-2618-2-4.
- Carlstein C., Søes L.M., Christensen J.J. Aerococcus christensenii as part of severe polymicrobial chorioamnionitis in a pregnant woman. Open Microbiol. J. 2016; 10: 27-31. https://dx.doi.org/10.2174/1874285801610010027.
- Nicholson A.C., Bell M., Humrighouse B.W., McQuiston J.R. Complete genome sequences for two strains of a novel fastidious, partially acid-fast, gram-positive Corynebacterineae bacterium, derived from human clinical samples. Genome Announc. 2015; 3(6): e01462-15. https://dx.doi.org/10.1128/ genomeA.01462-15.
- Bell M.E., Bernard K.A., Harrington S.M., Patel N.B., Tucker T.A., Metcalfe M.G. et al. Lawsonella clevelandensis gen. nov., sp. nov., a new member of the suborder Corynebacterineae isolated from human abscesses. Int. J. Syst. Evol. Microbiol. 2016; 66(8): 2929-35. https://dx.doi.org/10.1099 / ijsem.0.001122.
- Hiippala K., Kainulainen V., Kalliomäki M., Arkkila P., Satokari R. Mucosal prevalence and interactions with the epithelium indicate commensalism of Sutterella spp. Front. Microbiol. 2016; 7: 1706. https://dx.doi.org/10.3389/ fmicb.2016.01706.
- Balle C., Lennard K., Dabee S., Barnabas S.L., Jaumdally S.Z., Gasper M.A. et al. Endocervical and vaginal microbiota in South African adolescents with asymptomatic Chlamydia trachomatis infection. Sci. Rep. 2018; 8(1): 11109. https://dx.doi.org/10.1038/s41598-018-29320-x.
- Подгорная А.В., Maхмутходжаев А.Ш., Михеенко Г.А. Микрофлора влагалища при бактериальном вагинозе у беременных женщин. Современные проблемы науки и образования. 2015; 6; 212. [Podgornaya A.V., Makhmutkhodzhaev A.Sh., Mikheenko G.A. Vaginal microflora in pregnant women with a bacterial vaginosis. Modern problems of science and education. 2015; 6; 212. (in Russian)]. http://www.science-education.ru/ru/article/view?id=23589 (22.04.2020).
- Amabebe E., Chapman D.R., Stern V.L., Stafford G., Anumba D.O.C. Mid-gestational changes in cervicovaginal fluid cytokine levels in asymptomatic pregnant women are predictive markers of inflammation-associated spontaneous preterm birth. J. Reprod. Immunol. 2018; 126: 1-10. https://dx.doi.org/10.1016/j.jri.2018.01.001.
- Zhu L., Luo F., Hu W., Han Y., Wang Y., Zheng H. et al. Bacterial communities in the womb during healthy pregnancy. Front. Microbiol. 2018; 9: 2163. https://dx.doi.org/10.3389/fmicb.2018.02163.
Received 25.08.2020
Accepted 09.11.2020
About the Authors
Natalya B. Kuznetsova, Dr. Med. Sci., Professor at the Center for Simulation Training, RostSMU, Ministry of Health of Russia; Chief Physician of the Clinic of Professor Bushtyreva LLC. Tel.: +7(928)770-97-62. E-mail: lauranb@inbox.ru.344022, Russia, Rostov-on-Don, Nakhichevanskiy str., 29; 344011, Russia, Rostov-on-Don, Sobornyi str., 58/7.
Irina O. Bushtyreva, Dr. Med. Sci., Professor, Director of the Clinic of Professor Bushtyreva LLC. Tel.: +7(928)296-15-97. E-mail: kio4@mail.ru.
344011, Russia, Rostov-on-Don, Sobornyi str., 58/7.
Violetta S. Dybova, Ph.D. Student at RostSMU, Ministry of Health of Russia; obstetrician-gynecologist, Perinatal Center, Rostov-on-Don. Tel.: +7(961)272-04-12.
E-mail: viola-kovaleva@mail.ru. ORCID: 0000-0002-2391-8952.
344022, Russia, Rostov-on-Don, Nakhichevanskiy str., 29; 344068, Russia, Rostov-on-Don, Bodraya str., 90.
Viktoriya V. Barinova, Ph.D., Teaching Assistant at the Department of Obstetrics and Gynecology No. 1, RostSMU, Ministry of Health of Russia. Tel.: +7(928)909-55-68. E-mail: victoria-barinova@yandex.ru. 344022, Russia, Rostov-on-Don, Nakhichevanskiy str., 29.
Dmitrii E. Polev, Ph.D. (Bio.Sci.in genetics), Chief Biologist of the Medical and Genetic Center "Serbalab". Tel.: +7(911)256-05-23. E-mail: vdudurich@cerbalab.ru.
199106, Russia, St. Petersburg, V.O., Bolshoy ave., 90-2.
Mikhail V. Aseev, General Director of the Medical Genetics Center "Serbalab", Senior Researcher, Specialist in Laboratory Genetics
Tel.: +7(914)542-20-10. E-mail: vdudurich@cerbalab.ru. 199106, Russia, St. Petersburg, V.O., Bolshoy ave., 90-2.
Vasilisa V. Dudurich, Biologist-Geneticist, Director for Development of the Medical Genetics Center "Serbalab". Tel.: +7(914)542-20-10. E-mail: vdudurich@cerbalab.ru.
199106, Russia, St. Petersburg, V.O., Bolshoy ave., 90-2.
For citation: Kuznetsova N.B., Bushtyreva I.O., Dybova V.S., Barinova V.V., Polev D.E., Aseev M.V., Dudurich V.V. Vaginal microbiome in pregnant women with preterm prelabor rupture of membranes at 22–28 weeks' gestation.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2021; 1: 94-102 (in Russian)
https://dx.doi.org/10.18565/aig.2021.1.94-102



