Methylation of the TLR2 and IGF2/H19 ICR genes in the placenta and blood plasma in preeclampsia
Objective. To study the methylation profile of genes in the placenta and plasma of patients with preeclampsia. Materials and methods. The study included two groups of pregnant women. The main group consisted of 26 women with preeclampsia, 12 of them had moderate preeclampsia and 14 women had severe preeclampsia. The control group consisted of 26 women with normal pregnancy. The methylation level of 22 genes was determined. The study was conducted using Methylation-Sensitive High Resolution Melting curve analysis (MS-HRM). Results. The study of methylation of IGF2/H19 imprinting control region (ICR) showed a statistically significant decrease in the relative level of methylation in the placenta and blood plasma in patients with moderate and severe preeclampsia (p<0.05). There was a statistically significant increase in the level of the TLR2 gene methylation in the placenta and blood plasma in patients with severe preeclampsia in comparison with the patients with normal pregnancy (p<0.05). Conclusion. The results of the study indicate that the TLR2 gene methylation and the IGF2/H19 ICR influence the development of a systemic inflammatory response in preeclampsia. The level of gene methylation in blood plasma correlates with the level of genes in the placenta and can be used as early non-invasive markers of preeclampsia.Boris D.A., Krasnyi A.M., Kurevlev S.V., Sadekova A.A., Kan N.E., Tyutyunnik V.L.
Keywords
Preeclampsia is one of the leading causes of maternal and neonatal morbidity and mortality worldwide, affecting 2–8% of all pregnancies [1]. The etiology of this condition still remains unclear. Preeclampsia is known to be a multifactorial complication of pregnancy; its onset and progression may be due to the genetic background of the mother, immune maladaptation, abnormal infiltration of trophoblast into the spiral arteries of the uterus, endothelial cell dysfunction, as well as some extragenital diseases of the mother [2]. Although the molecular mechanisms that contribute to the development of preeclampsia are not fully understood, it is believed that the placenta plays a key role in the development of this complication [3]. Numerous studies show that the placenta of women with preeclampsia changes the expression of various genes [4, 5], which are part of the signaling pathways that regulate trophoblast invasion, angiogenesis, cell adhesion and survival, the renin-angiotensin-aldosterone system, and the immune response [6–8]. Altered gene expression in the placenta of women with preeclampsia may indicate that epigenetic modifications that regulate gene expression without changing the main DNA sequence play a certain role in its development. DNA methylation is the most studied epigenetic modification that occurs in CpG dinucleotides and is associated with transcriptional silence. Several studies have examined the role of epigenetic modifications, in particular DNA methylation in the placenta of pregnant women with preeclampsia. These studies have shown that a number of gene regions are hypermethylated and/ or hypomethylated in the placenta in preeclampsia in comparison with the control samples [9–11].
The aim of the research is to study the methylation profile of genes in the placenta and plasma of patients with preeclampsia.
Materials and Methods
In the present study, 52 placenta samples of women after spontaneous and operative delivery and 52 plasma samples of the same women obtained before delivery were studied. All women taking part in this study were treated and delivered in the National Medical Research Centre for Obstetrics, Gynecology and Perinatology, Moscow, Russia. The study included two groups of pregnant women. The main group consisted of 26 women with preeclampsia, 12 of them had moderate preeclampsia (MPE) and 14 women had severe preeclampsia (SPE). The control group consisted of 26 women with normal pregnancy. The study was approved by the Local Ethics Committee. All women were informed about the purpose of the study and signed an informed consent to participate in it. The criteria for inclusion in the study were singleton pregnancy at 22 to 40 weeks’ gestation, the age of pregnant women from 18 to 45 years, the absence of severe extragenital pathology, the presence of MPE or SPE (for the main group). The exclusion criteria were multiple pregnancies, acute infections, autoimmune and oncological diseases, pregnancy following assisted reproductive technologies, and severe extragenital pathology.
During the first stage of the study, genes that could play a potential role in the pathogenesis of preeclampsia were selected, namely HLAG, GNA12, VEGF, TIMP2, MMP2, DAPK3, LEP, SOCS2, MEST, DKK3, TLR2, BMP6, RASSF1, CEBPA, CDO1, DFNA5, CTCF, PITX2, PTEN, SEPT9, CDH1, IGF2/H19 imprinting control region (ICR). In the second part of the research, we studied the level of methylated genes in blood plasma that have aberrant methylation in the placenta of women with preeclampsia for determining their potential diagnostic and prognostic significance.
Peripheral blood plasma of pregnant women was collected before the onset of labor or surgery in amount of 5 ml in vacuum tubes containing ethylenediamine tetra acetic acid (EDTA), which were processed within an hour after sampling. The plasma was isolated by centrifugation in two stages at 4°C: the first – 10 min, 200 g, the second – 10 min, 4500 g. Plasma samples were stored at a temperature of -80°C. Chorion villi at the place of attachment of the umbilical cord were used in the study. DNA was isolated from the tissue using K-Sorb spin columns (Syntol, Russia) according to the manufacturer’s instructions. Then bisulfate conversion of DNA was performed using the kit EpiJET (Thermo Scientific, USA); as a result, all non-methylated cytosines were converted to uracils, and methylated cytosines remained unmodified. After that, a polymerase chain reaction (PCR) with primers was performed to a fragment of island methylation of candidate genes, which resulted in the replacement of uracils for thymines. Primers were selected due to the absence of CpG dinucleotides. The methylation level of the examined fragment was determined using Methylation-Sensitive High-Resolution Melting curve analysis (MS-HRM) with the help of Precision Melt Analysis Software, version 3 (Bio Rad, USA). For the quantitative assessment of methylation, relative fluoresce units (RFU) were used at the temperature of maximum divergence of curves according to the method described by A.M. Krasny et al. [12]. PCR and Methylation-Sensitive High-Resolution Melting curve analysis were performed using the CFX-96 amplifier (Bio Rad, USA).
Statistical analysis
The results are presented as the median, upper and lower quartiles of Me (Q1; Q3). The statistical significance of differences was determined using the nonparametric Mann-Whitney U test. The Kruskal- Wallis test by ranks was used for the comparison of three independent groups. Qualitative characteristics of the groups were compared using Fischer’s exact test. The differences were considered statistically significant at p<0.05. When comparing three groups, the correction for multiple comparisons was applied. The programs Attestat (Russia), Statistica 10 and OriginPro 8.5 (USA) were used for statistical processing of results and plotting.
Results
When analyzing the age of pregnant women in the group with SPE, women of the older age group were significantly more common (p=0.012) in comparison with the control group. No statistically significant differences were revealed between the patients with MPE and normal pregnancy as well as between the patients with MPE and SPE. The analysis of weight- height parameters did not identify any statistically significant differences. However, the average weight of patients was higher in the groups with MPE and SPE. When analyzing the diseases of the patients, it was found out that women with hereditary thrombophilia were significantly more common in the group with MPE (p<0.001). No significant differences in gynecological diseases were found among patients. Gestational age at delivery in patients with MPE and SPE was significantly lower in comparison with the control group (p<0.001), due to the inclusion criteria in the study groups.
The clinical characteristics of the examined pregnant women are presented in Table.
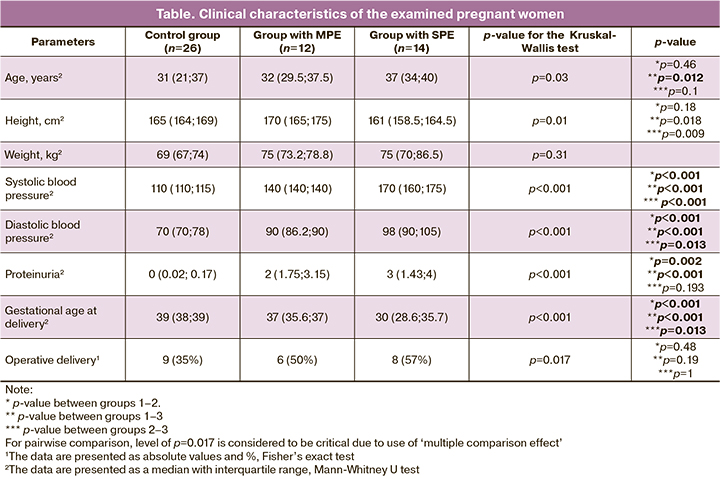
The analysis of methylation of 22 genes in the placenta revealed statically significant differences only in the TLR2 gene and IGF2/H19 ICR (Figure 1). The TLR2 gene showed an increased level of methylation only in patients with SPE (p=0.003). IGF2/H19 ICR showed a decrease in methylation, both in patients with MPE and SPE (p<0.001).
In order to determine the potential diagnostic and prognostic significance of the genes, we studied only the level of methylation of genes with aberrant methylation in the placenta. The level of the TLR2 and IGF2/H19 ICR in the blood plasma of pregnant women who participated in the study was analyzed. The results are shown in Figure 2.
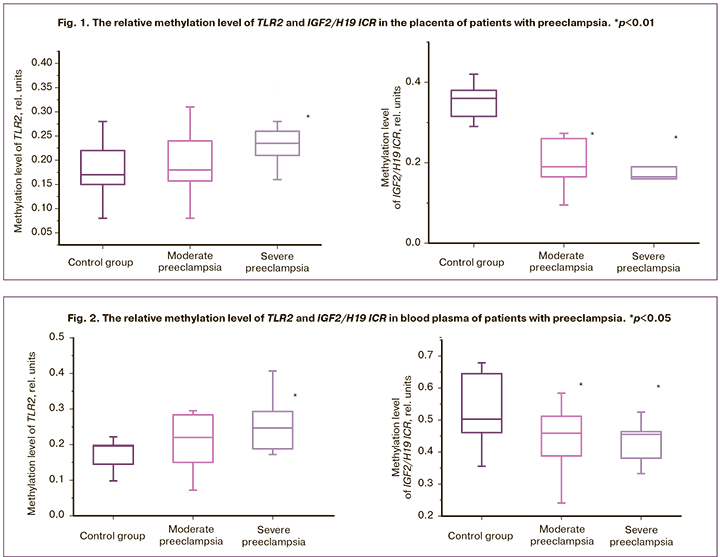
Significant differences in the TLR2 gene were revealed only in women with SPE (p=0.023). IGF2/H19 ICR had statistically significant differences both in patients with MPE (p=0.043) and SPE (p=0.038).
Discussion
It is known that the impairment of the formation and functioning of the placenta causes various obstetric complications [13, 14]. The developing pathological changes are characterized by the impairment of gene expression, which may result from aberrant DNA methylation. In this study, the profile of gene methylation in the placenta of patients with preeclampsia was analyzed. It was found that the TLR2 gene and the IGF2/H19 ICR had statistically significant differences in preeclampsia.
IGF2/H19 ICR regulates the expression of the imprinted IGF2 and H19 genes. A number of publications have shown a decrease in IGF2/H19 ICR methylation during fetal growth retardation [15, 16]. However, there are no findings on the decrease in methylation of this region in preeclampsia. Patients with preeclampsia showed a loss of imprinting in the placenta with an increase in the expression of the H19 gene [17], which may be associated with demethylation of IGF2/H19 ICR. It is known that H19 mRNA in cells performs an anti-inflammatory and anti-apoptotic function [18]; this fact suggests that a decrease in IGF2/H19 ICR methylation occurs as a response to increased activity of the immune system in preeclampsia for the adaptation of placental cells. Our studies have previously shown that in preeclampsia there is an increased activity of the immune system, leading to apoptosis in the placenta and an increase in the content of fetal DNA in mother’s blood [19–21].
When studying the TLR2 gene, we obtained the results suggesting an increase in the TLR2 gene methylation in patients with SPE, which may be indicative of its lower expression in the placenta. Being an important link in the immune defense, TLR2 is known to recognize various bacterial and viral pathogens. Therefore, it can be assumed that the impairment of this gene expression may cause incorrect recognition of infectious pathogens and the increase in their number in the placenta of patients with SPE. The described changes can play an important role in the development of pathological immune response and systemic inflammatory response syndrome which are characteristic of SPE. Some researchers notice the role of infectious factors in the pathogenesis of preeclampsia. For instance, F. Arechavaleta-Velasco et al. [22] showed that SPE is associated with the presence of adeno-associated virus 2 in the placenta. There is also evidence that the presence of single nucleotide polymorphisms (SNP) in the TLR2 gene in women may be associated with the development of preeclampsia [23]. W. Wujcicka et al. [24] found a relationship between the presence of SNP in the TLR2 gene and increased cytomegalovirus viral load in the fetus and mother. Thus, the role of the TLR2 gene in the pathogenesis of preeclampsia may be important, but the obtained results need further studies to be conducted.
Conclusion
Thus, we have obtained results that may be suggestive of the role of TLR2 gene methylation and IGF2/H19 ICR in the development of preeclampsia. It was found that the level of methylation of the examined genes in blood plasma reflects the level of methylation in the placenta. The above-mentioned data can be used to develop new methods for diagnosing and assessing the severity of preeclampsia.
References
- Pierik E., Prins J.R., van Goor H., Dekker G.A., Daha M.R., Seelen M.A.J., Scherjon S.A. Dysregulation of сomplement activation and placental dysfunction: a potential target to treat preeclampsia? Front. Immunol. 2020; 10: 3098. https://dx.doi.org/10.3389/fimmu.2019.03098.
- Lain K.Y., Roberts J.M. Contemporary concepts of the pathogenesis and management of preeclampsia. JAMA. 2002; 287(24): 3183-6.
- Myatt L., Muralimanoharan S., Maloyan A. Effect of preeclampsia on placental function: influence of sexual dimorphism, microRNA’s and mitochondria. Adv. Exp. Med. Biol. 2014; 814: 133-46. https://dx.doi.org/10.1007/978-1-4939-1031-1_12.
- Founds S.A., Conley Y.P., Lyons-Weiler J.F., Jeyabalan A., Hogge W.A., Conrad K.P. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta. 2009; 30(1): 15-24. https://dx.doi.org/10.1016/j.placenta.2008.09.015.
- Sitras V., Paulssen R.H., Grønaas H., Leirvik J., Hanssen T.A., Vårtun A., Acharya G. Differential placental gene expression in severe preeclampsia. Placenta. 2009; 30(5): 424-33. https://dx.doi.org/10.1016/j.placenta.2009.01.012.
- Cui Y., Wang W., Dong N., Lou J., Srinivasan D.K., Cheng W. et al. Role of corin in trophoblast invasion and uterine spiral artery remodelling in pregnancy. Nature. 2012; 484(7393): 246-50. https://dx.doi.org/10.1038/nature10897.
- Meng T., Chen H., Sun M., Wang H., Zhao G., Wang X. Identification of differential gene expression profiles in placentas from preeclamptic pregnancies versus normal pregnancies by DNA microarrays. OMICS. 2012; 16(6): 301-11. https://dx.doi.org/10.1089/omi.2011.0066.
- Sundrani D.P., Reddy U.S., Joshi A.A., Mehendale S.S., Chavan-Gautam P.M., Hardikar A.A. et al. Differential placental methylation and expression of VEGF, FLT-1 and KDR genes in human term and preterm preeclampsia. Clin. Epigenetics. 2013; 5(1): 6. https://dx.doi.org/10.1186/1868-7083-5-6.
- Yuen R.K., Peñaherrera M.S., von Dadelszen P., McFadden D.E., Robinson W.P. DNA methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur. J. Hum. Genet. 2010; 18(9): 1006-12. https://dx.doi.org/10.1038/ejhg.2010.63.
- Anton L., Brown A.G., Bartolomei M.S., Elovitz M.A. Differential methylation of genes associated with cell adhesion in preeclamptic placentas. PLoS One. 2014; 9(6): e100148. https://dx.doi.org/10.1371/journal.pone.0100148.
- Yeung K.R., Chiu C.L., Pidsley R., Makris A., Hennessy A., Lind J.M. DNA methylation profiles in preeclampsia and healthy control placentas. Am J Physiol. Heart Circ. Physiol. 2016; 310(10): H1295-303. https://dx.doi.org/10.1152/ajpheart.00958.2015.
- Красный А.М., Садекова А.А., Волгина Н.Е., Машаева Р.И., Кометова В.В., Хабас Г.Н., Голицына Ю.С., Носова Ю.В., Оводенко Д.Л. Исследование уровня метилирования гена RASSF1 в плазме и опухоли при раке эндометрия. Бюллетень экспериментальной биологии и медицины. 2019; 167(2): 223-7. [Krasnyi A.M., Sadekova A.A., Volgina N.E., Mashaeva R.I., Kometova V.V., Habas G.N., Golitsyna Yu.S., Nosova Yu.V., Ovodenko D.L. Study of the methylation of RASSF1 gene in plasma and tumor at endometrial cancer. Bulletin of experimental biology and medicine. 2019; 167 (2): 223-7. (in Russian)].
- Zárate A., Saucedo R., Valencia J., Manuel L., Hernández M. Early disturbed placental ischemia and hypoxia creates immune alteration and vascular disorder causing preeclampsia. Arch. Med. Res. 2014; 45(7): 519-24. https://dx.doi.org/10.1016/j.arcmed.2014.10.003.
- Burrows T.D., King A., Loke Y.W. Expression of adhesion molecules by endovascular trophoblast and decidual endothelial cells: implications for vascular invasion during implantation. Placenta. 1994; 15(1): 21-33.
- Хачатрян З.В., Кан Н.Е., Красный А.М., Садекова А.А., Куревлев С.В., Тютюнник В.Л. Метилирование генов в плаценте при задержке роста плода. Акушерство и гинекология. 2019; 12: 52-6. [Khachatryan Z.V., Kan N.E., Krasnyi A.M., Sadekova A.A., Kurevlev S.V., Tyutyunnik V.L. Methylation of genes in the placenta with fetal growth retardation. Akusherstvo i ginekologiya / Obstetrics and Gynecology, 2019; 12: 52-6. https://dx.doi.org/10.18565/aig.2019.12.54-58. (in Russian)]
- Koukoura O., Sifakis S., Soufla G., Zaravinos A., Apostolidou S., Jones A. et al. Loss of imprinting and aberrant methylation of IGF2 in placentas from pregnancies complicated with fetal growth restriction. Int. J. Mol. Med. 2011; 28(4): 481-7. https://dx.doi.org/10.3892/ijmm.2011.754.
- Yu L., Chen M., Zhao D., Yi P., Lu L., Han J. et al. The H19 gene imprinting in normal pregnancy and pre-eclampsia. Placenta. 2009; 30(5): 443-7. https://dx.doi.org/10.1016/j.placenta.2009.02.011.
- Li X., Wang H., Yao B., Xu W., Chen J., Zhou X. lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci. Rep. 2016; 6: 36340. https://dx.doi.org/10.1038/srep36340.
- Борис Д.А., Волгина Н.Е., Красный А.М., Тютюнник В.Л., Кан Н.Е. Прогнозирование преэклампсии по содержанию CD16-негагивных моноцитов. Акушерство и гинекология. 2019. 7. 49-55. [Boris D.A., Volgina N.E., Krasnyi A.M., Tyutyunnik V.L., Kan N.E. Prediction of preeclampsia on the couts of cd-16 negative monocytes. Akusherstvo i ginekologiya / Obstetrics and Gynecology. 2019; 7: 49-55. (in Russian)]. https://dx.doi.org/10.18565/aig.2019.7.49-55.
- Красный А.М., Грачева М.И., Садекова А.А., Вторушина В.В., Балашов И.С., Кан Н.Е., Боровиков П.И., Кречетова Л.В., Тютюнник В.Л. Комбинированное исследование общей, фетальной ДНК, цитокинов в плазме крови матери при преэклампсии. Бюллетень экспериментальной биологии и медицины. 2017; 164(12): 686-91. [Krasnyi A.M., Gracheva M.I., Sadekova A.A., Vtorushina V.V., Balashov I.S., Kan N.E., Borovikov P.I., Krechetova L.V., Tyutyunnik V.L., Complex Analysis of Total and Fetal DNA and Cytokines in Blood Plasma of Pregnant Women with Preeclampsia. Bulletin of experimental biology and medicine. 2017; 12: 686-91. (in Russian)]. https://dx.doi.org/10.1007/s10517-018-4066-1.
- Сухих Г.Т., Красный А.М., Кан Н.Е., Майорова Т.Д., Тютюнник В.Л., Ховхаева П.А., Сергунина О.А., Тютюнник Н.В., Грачева М.И., Вавина О.В., Озернюк Н.Д., Борис Д.А. Апоптоз и экспрессия ферментов антиоксидантной защиты в плаценте при преэклампсии. Акушерство и гинекология. 2015; 3: 11-5. [Sukhikh G.T., Krasnyj A.M., Kan N.E., Majorova T.D., Tyutyunnik V.L., Khovkhaeva P.A., Sergunina O.A., Tyutyunnik N.V., Gracheva M.I., Vavina O.V.,Ozerniuk N.D., Boris D.A. Placental apoptosis and antioxidant defense enzyme gene expression in preeclampsia. Akusherstvo i ginekologiya / Obstetrics and Gynecology, 2015; 3: 11-5. (in Russian)].
- Arechavaleta-Velasco F., Ma Y., Zhang J., McGrath C.M., Parry S. Adeno-associated virus-2 (AAV-2) causes trophoblast dysfunction, and placental AAV-2 infection is associated with preeclampsia. Am. J. Pathol. 2006; 168(6): 1951-9.
- Xie F., Hu Y., Speert D.P., Turvey S.E., Peng G., Money D.M. et al.; Toxaemia Study Group. Toll-like receptor gene polymorphisms and preeclampsia risk: a case-control study and data synthesis. Hypertens. Pregnancy. 2010; 29(4): 390-8. https://dx.doi.org/10.3109/10641950903242659.
- Wujcicka W., Paradowska E., Studzińska M., Wilczyński J., Nowakowska D. TLR2 2258 G>A single nucleotide polymorphism and the risk of congenital infection with human cytomegalovirus. Virol. J. 2017; 14(1): 12. https://dx.doi.org/10.1186/s12985-016-0679-z.
Received 01.03.2020
Accepted 27.03.2020
About the Authors
Daiana A. Boris, postgraduate student of the National Medical Research Center for Obstetrics, Gynecology and Perinatology named after Academician V.I. Kulakov Ministry of Health of Russia. Tel.: +7(91)081-89-97. E-mail: dayana_boris@mail.ru. ORCID ID 0000-0002-0387-4040.4, Ac. Oparina str., Moscow, 117997, Russian Federation.
Aleksey M. Krasnyi, PhD, the Head of the cytology laboratory of the National Medical Research Center for Obstetrics, Gynecology and Perinatology named after Academician V.I. Kulakov Ministry of Health of Russia. Tel.: +7(495)438-22-72. E-mail: alexred@list.ru.
4, Ac. Oparina str., Moscow, 117997, Russian Federation.
Sergey V. Kurevlev, scientific researcher of the cytology laboratory of the National Medical Research Center for Obstetrics, Gynecology and Perinatology named after Academician V.I. Kulakov Ministry of Health of Russia. Tel.: +7(985)693-66-33. E-mail: s_kurevlev@oparina4.ru.
4, Ac. Oparina str., Moscow, 117997, Russian Federation.
Alsu A. Sadekova, PhD, scientific researcher of the cytology laboratory of the National Medical Research Center for Obstetrics, Gynecology and Perinatology named after Academician V.I. Kulakov Ministry of Health of Russia. Tel.: +7(495)438-22-72. E-mail: a_sadekova@oparina4.ru.
4, Ac. Oparina str., Moscow, 117997, Russian Federation.
Natalia E. Kan, PhD, MD, head doctor of the Perinatal Center European Medical Center; Professor of the Department of Obstetrics and Gynecology of the National Medical Research Center for Obstetrics, Gynecology and Perinatology named after Academician V.I. Kulakov Ministry of Health of Russia. Tel.: +7(926)220-86-55.
E-mail: kan-med@mail.ru. Researcher ID B-2370-2015. ORCID ID 0000-0001-5087-5946125040.
15/1, Pravda str., Moscow, 125040, Russian Federation; 4, Ac. Oparina str., Moscow, 117997, Russian Federation.
Victor L. Tyutyunnik, PhD, MD, deputy head doctor of the Perinatal Center European Medical Center. Tel.: +7(903)969-50-41. E-mail: tioutiounnik@mail.ru.
Researcher ID B-2364-2015. ORCID ID 0000-0002-5830-5099. 15/1, Pravda str., Moscow, 125040, Russian Federation.
For citation: Boris D.A., Krasnyi A.M., Kurevlev S.V., Sadekova A.A., Kan N.E., Tyutyunnik V.L. Methylation of the TLR2 and IGF2/H19 ICR genes in the placenta and blood plasma in preeclampsia.
Akusherstvo i Ginekologiya / Obstetrics and Gynecology. 2020; 7: 93-98 (in Russian)
http://dx.doi.org/10.18565/aig.2020.7.93-98



