Uterine microbiome and immunohistochemical markers of chronic endometritis in recurrent pregnancy loss
Objective: To identify the genera of microorganisms that inhabit endometrium of healthy women and patients with recurrent pregnancy loss (RPL), as well as the genera of microorganisms causing chronic endometritis (CE).Barinova V.V., Kuznetsova N.B., Bushtyreva I.O., Dudurich V.V., Shatalov A.E.
Materials and methods: The endometrial microbiome was examined in 14 women with RPL and 15 fertile healthy women by sequencing the next generation 16S rRNA gene. In addition, correlation between CE, CD138, CXCL13 markers and members of various endometrial bacterial genera was analyzed.
Results: In patients of both groups, the most abundant bacterial genus in the endometrial samples was Lactobacillus (30.3% in patients with RPL, 29.3% in fertile patients). Statistically significant differences between the groups were found only for the bacterial genera Brevibacillus and Corynebacterium 1. The mean relative abundance of Brevibacillus was higher in the healthy fertile women [0.11 (0;0.3)%] than in the women with RPL [0 (0;0)% (p=0.008)]. The relative abundance of Corynebacterium 1 was 0 (0;0)% and 0.07 (0;0.13)% (p=0.002), respectively. The marker CD138 was associated with the presence of the bacteria of genus Pseudorhodoferax. The presence of marker CXCL13 was associated with bacteria of genera Alistipes, Butyricimonas, Dialister, Leuconostoc, Neisseria, Parabacteroides, Phascolarctobacterium, Prevotella, Ruminococcaceae, Sutterella, Sphingobium, Subdoligranulum.
Conclusion: Simultaneous analysis of the endometrial microbiome and CE markers can optimize preconception care in women with high perinatal risk.
Keywords
Endometrial disorders remain the focus of research as a cause of repeated implantation failure in recurrent pregnancy loss (RPL) and after embryo transfer in IVF. RPL is a significant medical and social issue affecting the quality of life of women of reproductive age. It is a multifactorial polyetiological condition caused by a number of genetic, immunological, endocrine, coagulation disorders, etc. However, a significant contribution to RPL pathogenesis is made by endometrial disorders, including those pathogenic microorganisms that inhabit it, leading to the development of the pathological process.
The study of the endometrial microbial community profile has long been considered a difficult task. Traditional culture techniques can identify only 1–2% of the available microorganisms [1–3]. The advent of next-generation sequencing has overcome two cultural method disadvantages: the presence of unculturable microorganisms and their high genetic biodiversity [4]. All currently available data on the composition of the endometrial microbial landscape suggest its association with reproductive outcomes in assisted reproduction and with various gynecological conditions, such as chronic endometritis (CE), endometriosis, dysfunctional uterine bleeding, endometrial polyps, and endometrial cancer or hyperplasia [5–7]. However, causality is difficult to prove because the reproductive tract is a polymicrobial niche and it is not clear whether uterine dysbiosis is a cause or a consequence of pathology.
Recently, the concept of a sterile uterus has been highly debated [8]. If bacteria or their particles are present in the uterine cavity, their role, even before pregnancy, in maintaining uterine homeostasis cannot be overestimated and certainly deserves attention.
The endometrium is a unique tissue that undergoes monthly cyclic changes, manifested as menstruation, proliferation, secretion, and decidualization under the influence of ovarian steroid hormones. The endometrium contains a considerable number of immunocompetent cells, natural killer (NK-cells), macrophages, T-cells, and neutrophils, whose percentages and density of distribution vary during the cycle. Cycle-dependent changes in leukocyte subpopulations and their mediators are likely to play a crucial role in implantation processes. Conversely, antibody-producing B-lymphocytes and plasma cells are rarely found in endometrial tissue. Bacteria are known to affect the immune system [9]. If bacteria impact the local immune status in the endometrium even before pregnancy, they can therefore influence endometrial receptivity, as well as early placentation. Successful implantation of a fertilized egg requires both endometrial transformation and a complex interaction between the blastocyst and the transformed endometrium. These processes largely depend on the receptive function of the endometrium. If the endometrium does not undergo the necessary transformations to reach a highly receptive state during the menstrual cycle, implantation disorders and even infertility occur as a consequence. Since bacteria play a role in morphological changes, such as in mucosal cells, it can be assumed that they have a direct influence on the decidualization processes. At the same time, the resident microflora, which is normal for a given biotope, can provide protection against pathogenic microorganisms, thus contributing to the normal functioning of the endometrium.
To date, few studies have focused on the endometrial microbiome and fertility, especially related to CE markers [10, 11]. In addition to standard histological criteria of CE (lymphocytic infiltration of the functional and basal endometrial layers, presence of plasma cells, stromal fibrosis, sclerotic changes in the spiral arteries, focal basal hyperplasia, gland deformity), there is a “gold standard” immunohistochemical diagnosis of CE. The main markers of plasma cells, mature tissue B lymphocytes, is CD138, or syndecan-1, which is an intercellular matrix protein. The study of chemokine CXCL13, which is a selective B-lymphocyte chemoattractant and participates in the immigration of B-lymphocytes into the endometrium, is also promising.
It remains unclear which bacteria may trigger an inflammatory response in the endometrium except for absolute pathogens, and how to correct these dysbiotic conditions. In many ways, we can use the knowledge about the intestinal microbiota and ideas about its correction as an approach to the treatment of various gastrointestinal diseases and normalization of the local immune response, and extrapolate this knowledge to the treatment of endometrial dysbiosis.
The present study aimed to identify the genera of microorganisms that inhabit the endometrium of healthy women and patients with recurrent pregnancy loss, as well as the genera of microorganisms causing chronic endometritis.
Materials and methods
Twenty-nine women divided into 2 groups were included in the study. The small sample size was justified by external constraints and the need to carefully select patients with RPL, excluding all other causes of RPL except endometritis. All patients were managed at the Department of Obstetrics and Gynecology No. 1, RostSMU, Ministry of Health of Russia. Group 1 included 14 women aged 20 to 42 years with RPL (2 or more losses) at 12 weeks of gestation. Exclusion criteria were any pathology of the uterus, including leiomyomas, polyps, congenital malformations, and intrauterine adhesions, as well as the usage of a contraceptive intrauterine device in the previous 6 months, any systematic inflammatory disease or severe extragenital pathology, and the recent use of systemic antibiotic therapy. All patients underwent molecular karyotyping of abortion material to rule out genetic factors of RPL. All aborted embryos had a normal chromosome set (Genomed Medical and Genetics Center, Optima molecular karyotyping of aborted material). Furthermore, women with congenital and acquired thrombophilia, endocrine disorders, menstrual disorders, and sexually transmitted infections confirmed by polymerase chain reaction (PCR) were excluded from the study. The second group enrolled 15 healthy women aged 20 to 42 years without complaints, without a history of obstetric or gynecological problems, who had at least one natural birth in their family history. The exclusion criteria, in addition to the criteria for Group 1, also included surgical abortions, spontaneous miscarriage, missed miscarriage, dilation, and curettage in the postpartum period, and a history of cesarean section.
All participants signed an informed consent to participate in the study. The Research Ethics Committee of the Kuban SMU reviewed and approved the study.
Sampling
Endometrial samples were taken during the luteal phase of the menstrual cycle from day 22 to day 24. To minimize the risk of contaminating endometrial samples in the vagina, a catheter with a hard outer part and an internal soft part used in embryo transfer was inserted into the uterine cavity, avoiding contact with the vaginal wall [12, 13]. After visualizing the cervix using a vaginal speculum, cervical mucus was removed by a sterile swab soaked in a chlorhexidine solution. Then, the catheter for embryo transfer was carefully inserted into the cervical canal without touching the vaginal walls. At the same time, the soft inner part of the catheter was inserted into the outer one, thus avoiding bacterial contamination of the cervical canal. After entering the uterine cavity, the inner catheter was gently pulled out of the outer end to the uterine fundus and a sample was taken. After that, the inner catheter was again pulled back to the outer one and the entire system was removed from the cervical canal and vagina. Finally, the endometrial sample was placed in an Eppendorf tube with a mucolytic transport medium (Central Research Institute of Epidemiology of Rospotrebnadzor, Moscow, Russia); the medium was kept at +4°C until DNA extraction.
After microbiome sampling, only Group 1 women with a history of RPL underwent a standard Pipelle endometrial sampling using a Yunona Classic aspiration probe. The aspiration probe was gently inserted through the cervical canal into the uterine cavity up to the uterine fundus, the internal piston was pulled out to create negative pressure. The probe was slowly removed from the uterine cavity using rotary movements to obtain pieces of endometrial tissue through small openings located at the tip of the catheter. Endometrial tissue obtained by biopsy was placed in 10% neutral buffered formalin solution for fixation at room temperature and subsequently embedded in paraffin for histopathological examination. Histological examination and immunohistochemical tests were performed for the presence of CD138 and CXCL13 markers.
Total DNA was isolated using a Ribo-Prep kit (Central Research Institute of Epidemiology Rospotrebnadzor, Russia) according to the manufacturer's protocol.
16S rRNA gene fragment libraries were prepared using the Illumina 16S Metagenomic Sequencing Library Preparation protocol (Part #15044223, Rev. B). Five ng of total DNA was amplified for 25 cycles using primers to V3 and V4 sites of the 16S rRNA gene of bacteria recommended in the protocol and KAPA HiFi HotStart ReadyMix PCR mix (2X) (Roche Diagnostics, Switzerland). The DNA fragments were purified using paramagnetic AMPure XP beads (Beckman Coulter, USA). For index PCR, 5 ng of DNA was subjected to 8 cycles of amplification after the first round of amplification using birkodinated primers from the Nextera XT Index Kit (Illumina, USA) and KAPA HiFi HotStart ReadyMix (2X) PCR mix (Roche Diagnostics, Switzerland). The obtained libraries were purified using paramagnetic particles, pooled in an equimolar ratio, and sequenced on a MiSeq System (Illumina, USA) in 2*151 paired-end sequencing mode.
The obtained sequencing data were analyzed using the custom bioinformatic pipeline implemented in the R v.3.6 programming language (R Core Team, 2014) and Python. At the first stage of the pipeline, the primer sequences were removed from the beginning of the readings. Reads that did not contain primers were also removed. Then, the last 25 nucleotides of reads were removed as having low quality, and the obtained data were processed using the DADA2 protocol to detect exact sequence variants [14]. After determining the exact variants of the sequences, the forward and reverse readings were combined by concatenation, and the obtained sequences were used for Bayesian taxonomic classification [15], using the SILVA v132 reference database [16]. Species were identified using the DADA2 complete match algorithm using SILVA v132 database sequences preprocessed with custom scripts to match the analysis.
Statistical analysis
The distribution of continuous variables was tested for normality using the Shapiro–Wilk test. All variables, except for age, were not normally distributed (p<0.05). Therefore, they were reported as median and interquartile range Me (Q1; Q3). Bacterial content was estimated in fractions of the total microbiome. Categorical variables were presented as counts and percentages, n (%). The comparison of continuous variables was made with the nonparametric Mann–Whitney test. The differences were considered statistically significant at p<0.05. Calculations were performed in R (version 3.2, R Foundation for Statistical Computing, Vienna, Austria).
Results
The clinical and demographic characteristics of the patients in both groups are presented in Table 1. Patients in both groups were comparable in body mass index, age at menarche, duration of menstrual cycle, duration of menstruation, and age. These variables were normally distributed in both groups.
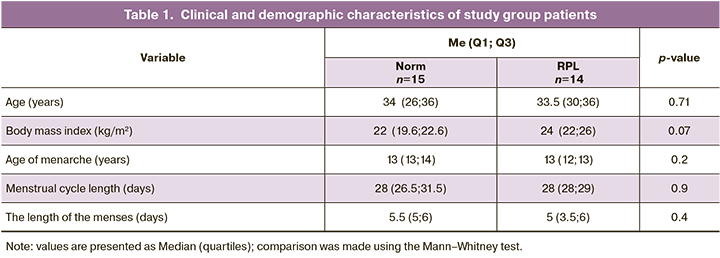
Next-generation sequencing analysis of the endometrial microbiome identified microorganisms belonging to a total of 19 phyla, 32 classes, 107 families, 241 genera, and 288 different species according to the taxonomic classification. The relative bacterial abundances from different genera in all patients is presented in Figure 1. Such a high biodiversity of genera and species is due, on the one hand, to the high accuracy of the sequencing technique and, on the other hand, to possible contamination during the collection of biomaterials and sequencing, as will be discussed below. Comparisons between groups were made at the taxonomic level.
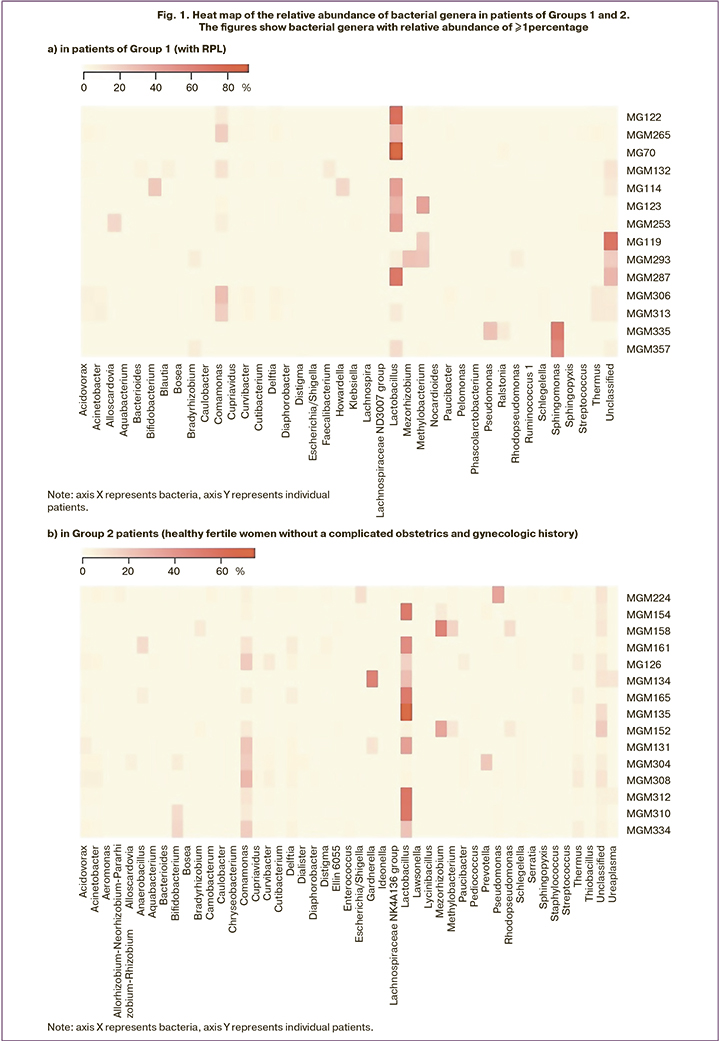
Analysis of the relative abundance of bacterial genera in the endometrial microbiome of all women in Groups 1 and 2 showed Lactobacillus spp. being the most abundant bacteria. The box plot (Fig. 2) of the relative abundance of Lactobacillus in the composition of the endometrial microbiota in groups 1 and 2 shows that there was no significant difference between the two groups; the medians in both groups were almost identical.
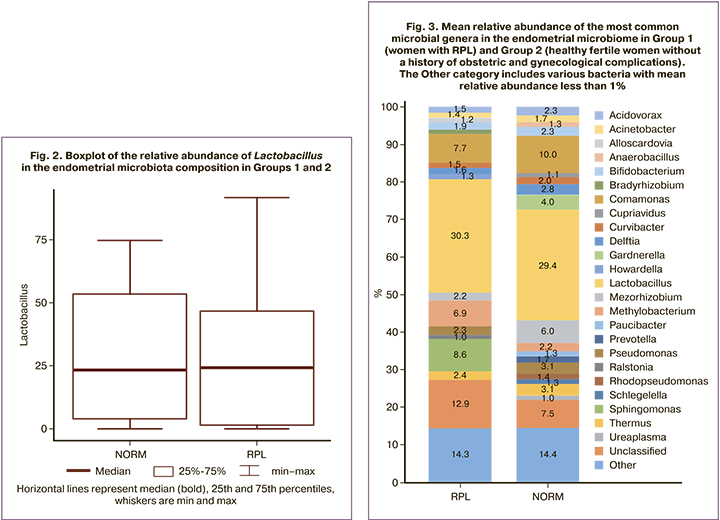
In women with RPL, the most abundant genera were Lactobacillus – 30.3%, Comamonas – 7.7%, Sphingomonas – 8.6%, in the group of healthy fertile patients, the most abundant genera were Lactobacillus – 29.4%, Comamonas – 16.8%, and Mesorhizobium – 6.0% (Fig. 3). A statistical comparison of the Lactobacilli proportions between the two groups revealed no statistically significant differences (median and interquartile intervals were 24 (1.48; 46.5)% and 23.3 (3.7; 53.4)%, respectively, p=0.99). In addition, there was no absolute dominance of lactobacilli in the composition of the uterine microbiome in the percentage of more than 90%, as shown in other studies [17].
Analysis of the relative abundance of microbial genera in the study groups showed statistically significant differences only in the relative abundance of bacteria of the genera Brevibacillus and Corynebacterium 1. In the group of healthy fertile women, the relative abundance of Brevibacillus was higher and was 0.11 (0; 0.3)%, in the group of women with RPL [0 (0; 0)% (p=0.008)]. In the group of healthy fertile women, the relative abundance of Corynebacterium 1 was lower and was 0 (0; 0)%, in the group of women with RPL [0.07 (0; 0.13)% (p=0.02)]. However, it cannot help noting that these genera of microorganisms, for which statistically significant differences were found, constituted less than 1% of the composition of the uterine microbiome compared to other microorganisms. Therefore, the differences between the groups have little clinical significance. Moreover, the presence of these bacteria can indirectly suggest contamination at any of the stages of material collection and sequencing.
Furthermore, endometrial biopsy specimens from patients with RPL were submitted for histological and immunohistochemical examination (markers CD138 and CXCL13) (Table 2).
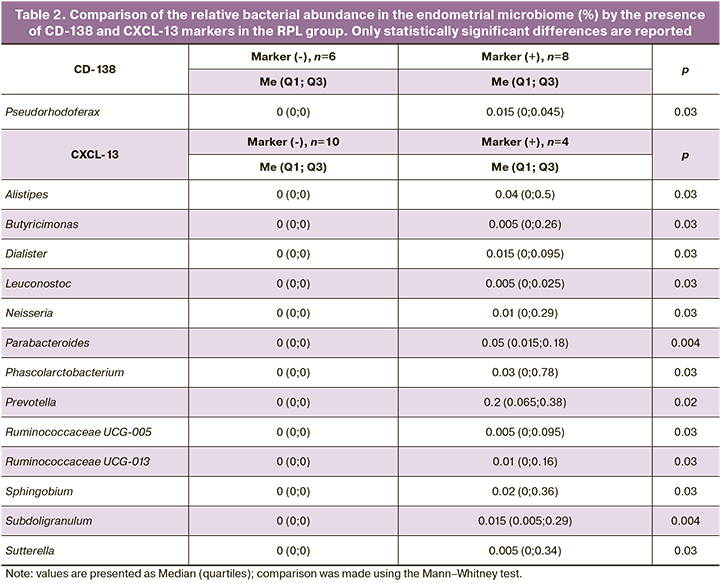
Plasma cell density was assessed and CE diagnosis was made when plasma cell density was greater than 5 per 10 mm2. Eight (57%) of 14 patients were diagnosed with CE according to this criterion. There was a positive association of the CE marker CD138 with a high relative abundance in the uterine microbiome of Pseudorhodoferax bacteria (0 (0; 0)% in women with a negative CD138 marker and 0.015 (0; 0.045)% with a positive marker, p=0.03). No statistically significant relationship was found for the other microbial genera. However, the relative abundance of this genus of bacteria was less than 1%, suggesting a questionable clinical significance of this correlation. However, for the bacteria of the genus Pseudorhodoferax, the human body is not a familiar habitat. Their natural habitat is soil, suggesting that their presence may be the result of contamination at any of the stages of the study.
The CXCL13 marker is not the gold standard for CE diagnosis, unlike the CD138 marker. However, as a selective B-lymphocyte chemoattractant, it can be used as a supplementary parameter for CE diagnosis. The CXCL13 marker was detected in four (29%) of all 14 women with RPL. Furthermore, these same four patients had a positive CD138 marker (that is, 4/14 patients with RPL had both CE markers).
The association of the CXCL13 marker with the presence of various bacterial genera in female patients was analyzed by comparing the relative abundance of these bacteria in the groups according to the presence of the marker. The presence of the CXCL13 marker was associated with an increased bacterial abundance of genera Alistipes (p=0.03), Butyricimonas (p=0.03), Dialister (p=0.03), Leuconostoc (p=0.03), Neisseria (p=0.03), Parabacteroides (p=0.004), Phascolarctobacterium (p=0.03), Prevotella (p=0.02), Ruminococcaceae UCG-005 (p=0.03), Ruminococcaceae UCG-013 (p=0.03), Sutterella (p=0.03), Sphingobium (p=0.03), and Subdoligranulum (p=0.004) (Table 2).
Discussion
Next-generation sequencing provides more accurate microbiome information than PCR because the genetic material of bacterial cells is amplified, revealing a greater diversity of the vaginal and endometrial microbiota in patients with CE. Molecular microbiology and metagenomics are excellent diagnostic tools to identify culturable and non-culturable endometrial pathogens in CE. New diagnostic techniques can shed light on the relationship between the chronic inflammatory process and new, understudied pathogens that are not detected by other diagnostic methods.
Before studying endometrial pathology, it is important to understand the normal endometrial microbiome. The data reported by different authors vary, and this difference depends on the uterine cavity sampling technique. Studies using transcervical sampling report higher rates of intrauterine lactobacilli colonization due to vaginal contamination of the endometrial specimens. For example, it has been shown that the dominance of lactobacilli in the uterine microbiome of over 90% is the key to successful implantation of a fertilized egg, successful pregnancy and live birth [17] in patients using assisted reproductive technology. However, according to other researchers who studied the endometrial microbiome during organ-removing surgery, the relative abundance of lactobacilli did not exceed 45% [18–21]. Furthermore, the biodiversity of the samples obtained by transcervical and transabdominal sampling differ; transcervical sampling produces a higher number of isolated bacteria. Therefore, our findings confirm the current concept of lactobacilli dominance in the composition of the endometrial microbiome.
It is still unclear whether the disrupted uterine microbiome is the cause or the consequence of gynecological disorders. Many studies investigating the endometrial microbiota have shown that it is equally important to identify pathogens causing a pathologic process and commensal microflora that contribute to homeostasis and health. An altered endometrial microbiome can serve as a predictor of disease and lead to implantation disorders and adverse reproductive outcomes [22–24].
Sequencing has obvious downsides – it is not entirely clear what to do with the study results, because sequencing does not answer the question about the sensitivity of the detected microbiota to a particular antibiotic therapy, as well as whether these are living microorganisms or just the genetic material of a long dead bacterium that is not clinically relevant. The simultaneous analysis of both endometrial microbiota by sequencing and CE markers allows us to bring this modern diagnostic modality closer to clinicians and use it as a promising new tool to prepare the endometrium for a successful pregnancy.
The study has a number of limitations. First, it has a small sample size. The 29 women in both groups certainly cannot be generalized to a large patient population, but such a small sample size can be explained by a careful approach to the group of women with RPL, an attempt to exclude other causes of RPL, except for an infectious factor. In addition, to objectify the sequencing results, it is important to have several control samples for each medium sample: operating room air, transport medium, etc., to exclude contamination with microorganisms not from the endometrium. The third limitation of the study still lies in the method of material collection, that is, transcervical access, which does not completely exclude possible contamination with cervical and vaginal microflora, despite the use of special two-way catheters and antiseptic treatment. The transabdominal method, which involves the collection of material after hysterectomy or uterine amputation, is considered to be better in terms of excluding possible contamination, but these operations never take place without absolute indications, including fibroids, endometrial cancer, etc. Furthermore, pathological conditions (polyposis, malignant processes) of the uterus can change the endometrial microbial composition.
The lactobacilli-dominated uterine flora is clearly an indicator of a healthy endometrium, conveying protection towards pathogenic species contributing to uterine health. According to our study, lactobacilli were dominant in both groups, but their relative abundance was less than 90%, as also shown by other authors [17]. Nevertheless, our data support the majority of studies that lactobacilli are the predominant genus in the endometrium.
In the group of women with RPL, the concentration of bacteria of the genus Brevibacillus was statistically significantly lower and the concentration of bacteria of the genus Corynebacterium 1 was higher. However, it should be noted that there is an extremely low relative abundance of the microorganism with statistically significant differences, i.e. less than 1% of the total biomass. Due to the low relative abundance of these microorganisms, one can hardly attribute clinical significance to the differences.
According to some studies [25–28], the number of bacterial DNA amplification cycles was almost identical to the number of amplification cycles in the microbiome of sterile water, operating room air or other sterile media, indicating possible contamination of the samples. Thus, when discussing the study by Aagaard K. on the presence of placental microbiome [29–31], we assume that the isolated bacteria of genera Thioalkalivibrio, Pseudoalteromonas, Gloebacter, Arthrobacter are normal inhabitants of soil, lake water, bogs and sea water, which can indirectly testify to contamination at any stage of analysis. The same can be said about our findings. Many bacterial genera found in the endometrial microbiome of our patients are characteristic of other ecological niches – soil, marshes, lakes, and industrial sites, indirectly confirms the possibility of sample contamination.
When assessing the association of the CE marker CD138 with the presence of certain genera of microorganisms, the relationship of the verified CE with the presence of bacteria of the genus Pseudorhodoferax was found. These bacteria are not characteristic of the human body; they are inhabitants of soils and water. In addition, their relative abundance is also low. It is difficult to reliably assert whether this is a sign of environmental contamination at any stage of material collection or sequencing or whether this bacterial genus is still associated with a chronic inflammatory process in the endometrium. As for the promising marker CXCL13, a plasma cell chemoattractant, its presence has been associated with many bacterial genera, including Alistipes, Butyricimonas, Dialister, Leuconostoc, Neisseria, Parabacteroides, Phascolarctobacterium, Prevotella, Ruminococcaceae, Sutterella, Sphingobium, Subdoligranulum. Many of them are known to clinicians as potential CE pathogens. However, the CXCL13 marker was verified in only 4 of 14 women with RPL. CD138, the gold standard for CE diagnosis, was also detected in these four women. This small sample size also limits clinical interpretation of the findings.
Conclusion
The concept of the non-sterile uterus and the feasibility of improving reproductive outcomes in infertile patients by investigating the endometrial microbiota are no longer in doubt. More than 280 different genera of microorganisms have been identified in the endometrial microbiome, which, on the one hand, certainly confirms the fact that this biotope is non-sterile; on the other hand, such a large number of isolated genera may indirectly indicate potential microbial contamination at any of the stages of study – from material collection to transportation and the sequencing. To exclude contamination, it is essential that endometrial microbiota studies include technical controls for the material of a particular patient, which increases the cost of the study due to the high cost of sequencing.
Our findings show that in both fertile and RPL patients, the dominant bacterial genus in the endometrium was Lactobacillus, which confirms the available publications. Their relative abundance did not differ significantly between the groups. If the relative abundance of lactobacilli was almost identical, logic dictates that the presence of other bacteria in the uterine microbiome may determine reproductive failure. However, reliable differences were found only for the bacterial genera Brevibacillus and Corynebacterium 1, whose relative abundance in the endometrium was less than 1%, which is crucial in the clinical interpretation of these data. It is difficult to assume that bacteria, which are so rare in the endometrium, can affect the processes of implantation of a fertilized egg and the further development of the embryo.
There is no doubt that the simultaneous study of uterine microbiome and immunohistochemical CE markers is clinically feasible and justified, but even at this stage the findings are quite controversial. The microbial genera associated with markers of chronic inflammatory process in utero were also found in rather low numbers. At the same time, these microbial genera are more often characteristic of other ecological niches of nature than of the human body, which may also indirectly indicate sample contamination and the extreme need for control specimens at all stages of the study.
A number of limitations of the present study cast doubt on its results and lend support for further sufficiently powered studies. However, the present work can be considered a pilot study that is potentially useful for further research in this area.
References
- Wade W. Unculturable bacteria--the uncharacterized organisms that cause oral infections. J. R. Soc. Med. 2002; 95(2): 81-3. https://dx.doi.org/10.1258/jrsm.95.2.81.
- Wilson M.J., Weightman A.J., Wade W.G. Applications of molecular ecology in the characterisation of uncultured microorganisms associated with human disease. Rev. Med. Microbiol. 1997; 8: 91-101.
- Oliver J.D. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 2010; 34(4): 415-25. https://dx.doi.org/10.1111/j.1574-6976.2009.00200.x.
- Giudice L.C. Challenging dogma: the endometrium has a microbiome with functional consequences! Am. J. Obstet. Gynecol. 2016; 215(6): 682-3. https://dx.doi.org/10.1016/j.ajog.2016.09.085.
- Кузнецова Н.Б., Буштырева И.О., Дыбова В.С., Баринова В.В., Полев Д.Е., Асеев М.В., Дудурич В.В. Микробиом влагалища у беременных с преждевременным разрывом плодных оболочек в сроке от 22 до 28 недель беременности. Акушерство и гинекология. 2021; 1: 94-102. [Kuznetsova N.B., Bushtyreva I.O., Dybova V.S., Barinova V.V., Polev D.E., Aseev M.V., Dudurich V.V. Vaginal microbiome in pregnant women with preterm prelabor rupture of membranes at 22–28 weeks' gestation. Obstetrics and Gynecology. 2021; 1: 94-102. (in Russian)]. https://dx.doi.org/10.18565/aig.2021.1.94-102.
- Баринова В.В., Кузнецова Н.Б., Буштырева И.О., Соколова К.М., Полев Д.Е., Дудурич В.В. Микробиом верхних отделов женской репродуктивной системы. Акушерство и гинекология. 2020; 3: 12-7. [Barinova V.V., Kuznetsova N.B., Bushtyreva I.O., Sokolova K.M., Polev D.E., Dudurich V.V. The microbiome of the upper female reproductive tract. Obstetrics and Gynecology. 2020; 3: 12-7. (in Russian)]. https://dx.doi.org/10.18565/aig.2020.3.12-17.
- Баринова В.В., Кузнецова Н.Б., Буштырева И.О., Оксенюк О.С., Дудурич В.В., Шаталов А.Е. Микробиом эндометрия при многократных неудачах вспомогательных репродуктивных технологий и у здоровых женщин: где норма и где патология? Акушерство и гинекология. 2021; 6: 105-14. [Barinova V.V., Kuznetsova N.B., Bushtyreva I.O., Oksenyuk O.S., Dudurich V.V., Shatalov A.E. Endometrial microbiome in women with and without a history of repeated failures of assisted reproductive technology: what are norm and pathology? Obstetrics and Gynecology. 2021; 6: 105-14. (in Russian)]. https://dx.doi.org/10.18565/aig.2021.6.105-114.
- Perez-Munoz M.E., Arrieta M.C., Ramer-Tait A.E., Walter J. A critical assessment of the ‘sterile womb’ and ‘in utero colonization’ hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017; 5: 48. https://dx.doi.org/10.1186/s40168-017-0268-4.
- Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012; 336(6086): 1268-73. https://dx.doi.org/10.1126/science.1223490.
- Кебурия Л.К., Смольникова В.Ю., Припутневич Т.В., Муравьева В.В., Трофимов Д.Ю., Шубина Е.С., Кочеткова Т.О. Микробиота полости матки и неудачи имплантации: есть ли связь? Акушерство и гинекология. 2021; 7: 133-43. [Keburiya L.K., Smol’nikova V.Yu., Priputnevich T.V., Murav’eva V.V., Trofimov D.Yu., Shubina E.S., Kochetkova T.O. Uterine microbiota and implantation failure: is there a link? Obstetrics and Gynecology. 2021; 7: 133-43. (in Russian)]. https://dx.doi.org/10.18565/aig.2021.7.133-143.
- Кебурия Л.К., Смольникова В.Ю., Припутневич Т.В., Муравьева В.В., Калинина Е.А. Микробиота эндометрия и репродуктивный исход в программах вспомогательных репродуктивных технологий. Акушерство и гинекология. 2020; 4: 166-72. [Keburia L.K., Smolnikova V.Yu., Priputnevich T.V., Muravyeva V.V., Kalinina E.A. Endometrial microbiota and reproductive outcome in assisted reproductive technology program. Obstetrics and Gynecology. 2020; 4: 166-72. (in Russian)]. https://dx.doi.org/10.18565/aig.2020.4.166-172.
- Verstraelen H., Vilchez-Vargas R., Desimpel F., Jauregui R., Vankeirsbilck N., Weyers S. et al. Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1-2 region of the 16S rRNA gene. PeerJ. 2016; 4: e1602. https://dx.doi.org/10.7717/peerj.1602.
- Liu Y., Ko E.Y., Wong K.K., Chen X., Cheung W.C., Law T.S. et al. Endometrial microbiota in infertile women with and without chronic endometritis as diagnosed using a quantitative and reference range-based method. Fertil. Steril. 2019; 112(4): 707-17.e1. https://dx.doi.org/10.1016/j.fertnstert.2019.05.015.
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016; 13(7): 581-3. https://dx.doi.org/10.1038/nmeth.3869.
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007; 73(16): 5261-7. https://dx.doi.org/10.1128/AEM.00062-07.
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013; 41(Database issue): D590-6. https://dx.doi.org/10.1093/nar/gks1219.
- Moreno I., Codoñer F.M., Vilella F., Valbuena D., Martinez-Blanch J.F., Jimenez-Almazán J. et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am. J. Obstet. Gynecol. 2016; 215(6): 684-703. https://dx.doi.org/10.1016/j.ajog.2016.09.075.
- Mitchell C.M., Haick A., Nkwopara E., Garcia R., Rendi M., Agnew K. et al. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am. J. Obstet. Gynecol. 2015; 212(5): 611.e1-9. https://dx.doi.org/10.1016/j.ajog.2014.11.043.
- Fang R.L., Chen L.X., Shu W.S., Yao S.Z., Wang S.W., Chen Y.Q. Barcoded sequencing reveals diverse intrauterine microbiomes in patients suffering with endometrial polyps. Am. J. Transl. Res. 2016; 8(3): 1581-92.
- Winters A.D., Romero R., Gervasi M.T., Gomez-Lopez N., Tran M.R., Garcia-Flores V. et al. Does the endometrial cavity have a molecular microbial signature? Sci. Rep. 2019; 9(1): 9905. https://dx.doi.org/10.1038/s41598-019-46173-0.
- Chen C., Song X., Wei W., Zhong H., Dai J., Lan Z. et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun. 2017; 8(1): 875. https://dx.doi.org/10.1038/s41467-017-00901-0.
- Baker J.M., Chase D.M., Herbst-Kralovetz M.M. Uterine microbiota: residents, tourists, or invaders? Front. Immunol. 2018; 9: 208. https://dx.doi.org/10.3389/fimmu.2018.00208.
- Moreno I., Codoñer F.M., Vilella F., Valbuena D., Martinez-Blanch J.F., Jimenez-Almazán J. et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am. J. Obstet. Gynecol. 2016; 215(6): 684-703. https://dx.doi.org/10.1016/j.ajog.2016.09.075.
- José B., Carlos S. Implantation failure of endometrial origin: what is new? Curr. Opin. Obstet. Gynecol. 2018; 30(4): 229-36. https://dx.doi.org/10.1097/GCO.0000000000000468.
- de Goffau M.C., Lager S., Sovio U., Gaccioli F., Cook E., Peacock S.J. et al. Human placenta has no microbiome but can contain potential pathogens. Nature. 2019; 572(7769): 329-34. https://dx.doi.org/10.1038/s41586-019-1451-5.
- Lager S., de Goffau M.C., Sovio U., Peacock S.J., Parkhill J., Charnock-Jones D.S., Smith G.C.S. Detecting eukaryotic microbiota with single-cell sensitivity in human tissue. Microbiome. 2018; 6(1): 151. https://dx.doi.org/10.1186/s40168-018-0529-x.
- Lauder A.P., Roche A.M., Sherrill-Mix S., Bailey A., Laughlin A.L., Bittinger K. et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 2016; 4(1): 29. https://dx.doi.org/10.1186/s40168-016-0172-3.
- Leiby JS., McCormick K., Sherrill-Mix S., Clarke E.L., Kessler L.R., Taylor L.J. et al. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome. 2018; 6(1): 196. https://dx.doi.org/10.1186/s40168-018-0575-4.
- Aagaard K., Ma J., Antony K.M., Ganu R., Petrosino J., Versalovic J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014; 6(237): 237ra65. https://dx.doi.org/10.1126/scitranslmed.3008599.
- Ходжаева З.С., Горина К.А., Тимошина И.В., Припутневич Т.В. Программирование здоровья новорожденного – роль материнского микробиома. Акушерство и гинекология: новости, мнения, обучение. 2019; 7(4): 61-5. [Khodzhaeva Z.S., Gorina K.A., Timoshina I.V., Priputnevich T.V. Infant health programming – the role of maternal microbiome. Obstetrics and Gynecology: News, Opinions, Training. 2019; 7(4): 61-5. (in Russian)]. https://dx.doi.org/10.24411/2303-9698-2019-14004.
- Riganelli L., Iebba V., Piccioni M., Illuminati I., Bonfiglio G., Neroni B. et al. Structural variations of vaginal and endometrial microbiota: hints on female infertility. Front. Cell. Infect. Microbiota. 2020; 10: 350. https://dx.doi.org/10.3389/fcimb.2020.00350.
Received 27.12.2021
Accepted 14.03.2022
About the Authors
Viktoriya V. Barinova, PhD, Teaching Assistant at the Department of Obstetrics and Gynecology No. 1, Rostov State Medical University, Ministry of Healthof the Russian Federation, 344022, Russia, Rostov-on-Don, str. Nakhichevanskiy, 29; Head of the Obstetric Department, Clinic of Professor Bushtyreva,
344011, Russia, Rostov-on-Don, str. Soborniy, 58/7, +7(928)909-55-68, victoria-barinova@yandex.ru, https://orcid.org/0000-0002-8584-7096,
WoS Researcher ID: https://publons.com/researcher/AAH-3314-2019, Scopus Author ID: https://www.scopus.com/authid/detail.uri?authorId=2578513
Natalya B. Kuznetsova, Dr. Med. Sci., Professor at the Center for Simulation Training, Rostov State Medical University, Ministry of Health of the Russian Federation,
344022, Russia, Rostov-on-Don, str. Nakhichevanskiy, 29; Chief Physiсian of the Clinic of Professor Bushtyreva, 344011, Russia, Rostov-on-Don, str. Sobornyi, 58/7, +7(928)770-97-62, lauranb@inbox.ru
Irina O. Bushtyreva, Dr. Med. Sci., Professor, Director of the Clinic of Professor Bushtyreva, 344011, Russia, Rostov-on-Don, str. Sobornyi, 58/7,
+7(928)296-15-97, kio4@mail.ru
Vasilisa V. Dudurich, Biologist-Geneticist, Director for Development, Medical Genetics Center Serbalab, 199106, Russia, St. Petersburg, V.O., Bolshoy ave., 90, build. 2, +7(914)542-20-10, vdudurich@cerbalab.ru
Alexander E. Shatalov, 1st year resident at the Department of Obstetrics, Gynecology and Perinatology of the FPK and PPS, Kuban State Medical University,
Ministry of Health of the Russian Federation, 350063, Russia, Krasnodar, M. Sedina str., 4, +7(928)191-35-82, shatal321@mail.ru
Authors' contributions: Barinova V.V. – conception and design of the study, manuscript drafting; Barinova V.V., Shatalov A.E., Dudurich V.V. – material collection and processing; Barinova V.V., Shatalov A.E. – statistical analysis; Bushtyreva I.O., Kuznetsova N.B. – manuscript editing.
Conflicts of interest: The authors have no conflicts of interest to declare.
Funding: This work was supported by grant no. 19-75-00006 "The role of the uterine cavity microbiome in the genesis of reproductive losses and ART failures" of the Russian Science Foundation.
Ethical Approval: The study was reviewed and approved by the Research Ethics Committee of the Kuban SMU (Rostov-on-Don, Russia).
Acknowledgment: The authors express their gratitude to the employees of the genetic laboratory of Serbalab LLC (St. Petersburg) Mikhail V. Aseev, Dmitry E. Polev, and Yuri A. Barbitov.
Patient Consent for Publication: All patients provided informed consent for the publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator Barinova V.V., Victoria-barinova@yandex.ru
For citation: Barinova V.V., Kuznetsova N.B., Bushtyreva I.O.,
Dudurich V.V., Shatalov A.E. Uterine microbiome and
immunohistochemical markers of chronic endometritis in recurrent pregnancy loss.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2022; 4: 84-94 (in Russian)
https://dx.doi.org/10.18565/aig.2022.4.84-94



