Exploration of the association between gonadotropin genes and their receptor gene variants and the development of polycystic ovary syndrome
Ustenko K.A., Derevyanchuk E.G., Aleksandrova A.A.
Objective: To evaluate the association between gonadotropin genes and their receptor gene variants with the development of polycystic ovary syndrome (PCOS).
Materials and methods: The study included 112 women. The main group comprised 57 women with PCOS, and the control group comprised 55 women. Exploration of gene variants NM_000894.3(LHB): c. 364G>A (p. Gly122Ser), rs5030774; NM_001382289. 1(FSHB):c.236_237del(p.Val79fs), rs5030646 and NC_000011.10:g.30204981G>A, rs11031006; NM_000233.4 (LHCGR):c.537-235G>A, rs4953616; NM_000145. 4(FSHR):c.919G>A(p.Ala307Thr), rs6165 was performed using allele-specific real-time PCR method followed by statistical data processing.
Results: Our study found no association between PCOS and the variant rs5030774 in the LHB gene and the variant rs5030646 in the FSHB gene. Statistically significant differences were found between the groups under study in distribution of alleles and genotypes across the variant rs11031006 in the FSHB gene, rs4953616 in the LHCGR gene, and rs6165 in the FSHR gene. Allele frequencies of the FSHB A rs11031006 and the FSHR T rs6165 were higher in the control group (59/110 (53.6%) at p=0.008 (OR=0.485 95%, CI 0.284–0.829) and 86/110 (78.2%) at p=0.014 (OR=0.478, 95%, CI 0.265–0.864), respectively, while the prevalence of the allele T rs4953616 of the LHCGR gene was higher in the group of patients with PCOS compared with the control group (87/114 (76,3%) at p=0.002 (OR=2.404, 95% CI 1.354–4.267)).
Conclusion: Exploration of polymorphic loci of gonadotropin genes and their receptor gene variants showed statistically significant association between PCOS and rs11031006 in the FSHB gene, rs4953616 in the LHCGR gene, and rs6165 in the FSHR gene.
Authors' contributions: Ustenko K.A. – collection and processing of primary material, genotyping; Derevyanchuk E.G. – analysis of literature data, statistical analysis of the obtained data, manuscript writing; Alexandrova A.A. – development of the concept and design of the study, manuscript editing.
Conflicts of interest: The authors confirm that they have no conflicts of interest to declare.
Funding: The study was supported by the Russian Science Foundation, project No. 23-15-00464.
Ethical Approval: The study was approved by the Ethics Committee of the Academy of Biology and Biotechnology named after D.I. Ivanovskiy of the Southern Federal University (Protocol No. 12 of December 24, 2019).
Patient Consent for Publication: All patients provided informed consent for the publication of their data (and associated images)
Authors' Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator.
For citation: Ustenko K.A., Derevyanchuk E.G., Aleksandrova A.A. Exploration of the association between
gonadotropin genes and their receptor gene variants and the development of polycystic ovary syndrome.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2025; (8): 143-149 (in Russian)
https://dx.doi.org/10.18565/aig.2025.123
Keywords
Polycystic ovary syndrome (PCOS) is a multifactorial and polygenic disorder affecting women of reproductive age. PCOS is characterized by oligoovulatory or anovulatory cycles, hirsutism, and polycystic ovaries. PCOS can be difficult to diagnose correctly due to a variety of signs and symptoms.
Manifestations of PCOS can be diverse. Some women have no symptoms, while others have many gynecological or metabolic disorders. At the same time, there are differences in the clinical picture depending on the ethnicity and place of residence. Impaired follicle development and reproductive problems in PCOS indicate that genetic variations affecting this process can play an important role in the disease development. In recent years, exploration of the genetic factors of PCOS has led to understanding of the multifaceted nature of this disease [1, 2]. Currently, advanced genetic methods are used to examine polymorphic regions of the genome associated with multifactorial diseases. As of April 2025, a collection of all published genome-wide association studies (the GWAS Catalog) includes 30 studies, which have identified 144 polymorphic loci associated with PCOS [3].
More than 50 genetic loci associated with PCOS were identified in women of European ethnicities. These include polymorphic loci of the THADA, DENND1A, and FSHB genes, as well as the FSHR and LHCGR genes. In PCOS, elevated serum levels of luteinizing hormone (LH), normal/low levels of follicle-stimulating hormone (FSH), and elevated ratio between LH and FSH were detected [4].
It was found that in different ethnic groups, polymorphic loci of the genes encoding FSH and LH, as well as their receptors, were associated with PCOS, that suggests the presence of a universal genetic profile predisposing to the development of PCOS in different populations [5–9].
The objective of the study was to evaluate the association between gonadotropin genes and their receptor gene variants with the development of polycystic ovary syndrome.
Material and methods
The study included 112 women aged 22–40 years, who received assisted reproductive technology treatment at the Center for Human Reproduction and IVF (Rostov-on-Don). Among the participants, 57 women were diagnosed with PCOS in accordance with the Rotterdam Criteria [10]. The inclusion criteria in the group of women with PCOS were at least two of three signs – the presence of clinical or biochemical evidence of hyperandrogenism, ovulatory disorders (oligoovulation or anovulation), and ovarian structure defined as polycystic ovaries. The control group consisted of 55 women, who underwent in vitro fertilization (IVF) due to tubal factor infertility or male infertility. The exclusion criteria for both groups comprised the diagnosis of endocrine disorders (for example, hyperprolactinemia or Cushing's disease) and ovarian neoplasms. The written informed consent to participate in the study was obtained from all patients. The study was conducted in accordance with ethical norms established by the Declaration of Helsinki, and was approved by the Ethics Committee of the Academy of Biology and Biotechnology named after D.I. Ivanovskiy of the Southern Federal University (Protocol No. 12 of December 24, 2019).
The patients in both groups were comparable in age and body mass index (BMI). Blood tests were performed to assess the endocrine status in the early follicular phase (days 3–5) of the menstrual cycle before starting hormonal ovarian stimulation in the programs of assisted reproductive technology. The competitive versions of solid-phase enzyme-linked immunosorbent assay were used for most of hormones (Alkor Bio, Russia, and DRG, Germany). Insulin-like growth factor 1 (IGF-1) was analyzed using solid phase sandwich enzyme immunoassay (DSL, USA).
In our study, DNA samples were obtained from the peripheral blood leukocytes using thermocoagulation technique and a set of reagents "DNA-Express-Blood".
The following gene variants were assessed: NM_000894.3(LHB):c. 364G>A (p.Gly122Ser), rs5030774; NM_001382289.1(FSHB):c. 236_237del (p.Val79fs), rs5030646 and NC_000011. 10:g.30204981G>A, rs11031006; NM_000233. 4(LHCGR):c.537-235G>A, rs4953616; NM_000145.4(FSHR):c.919G>A (p.Ala307Thr), rs6165.
The variant rs5030774 in the LHB gene was detected using real-time polymerase chain reaction (RTl-PCR) and the SNP-Screen kit (Syntol, Russia).
The developed primer pairs (Table 1) and reagents supplemented with SYBR Green I dye (Syntol, Russia) were used to analyze the variant rs4953616 in the LHCGR gene and the variant rs6165 in the FSHR gene using RT-PCR. The variant rs11031006 in the FSHB gene was detected using a set of primers (Table 1) and the 5X qPCRmix-HS SYBR reagents (Evrogen, Russia). The PCR program was selected experimentally, based on the manual [11] and recommendations of the manufacturer (Syntol, Russia) [11].
Statistical analysis
Statistical analysis was performed using Statistica 6.0 software. The Shapiro-Wilk test was used to check the data for normal distribution. The samples had non-normal distribution, for this reason, the Mann–Whitney U-test was used to identify statistical differences between the group of women with PCOS and the control group. The differences were considered to be statistically significant at p<0.05.
Compliance with the genotype frequency distribution to the Hardy–Weinberg equilibrium was checked using the online Hardy–Weinberg Equilibrium Calculator https://www.sebc.me/bioblog/labs/hwe-calculator. Compliance of genotype frequencies of the analyzed genes with the Hardy–Weinberg equilibrium was checked using Pearson’s chi-square (χ2) test. The differences in distribution of allelic variants in the genes in the groups under study were evaluated using Pearson’s chi-square (χ2) test and the BioStat software.
The odds ratio (OR) was calculated to estimate the risk of developing PCOS, using the formula OR=(A/B)×(C/D), where A and C are the number of women with and without PCOS, who have the mutant genotype, respectively; B and D are the number of women, who have no the mutant genotype in two groups. The OR is indicated with 95% confidence interval (CI). When 95% CI for OR did not include 1, the results were considered to be statistically significant [12].
Results and discussion
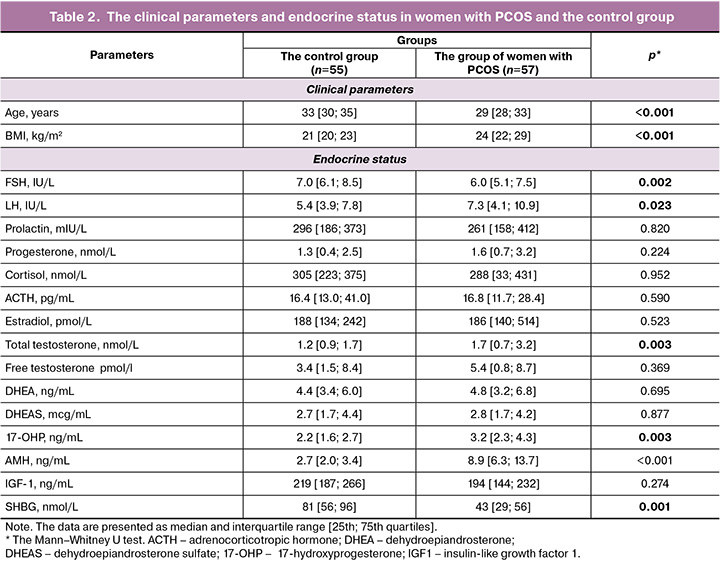
PCOS is characterized by hormonal imbalances. Serum hormone levels in the group of women with PCOS and the control group are shown in Table 2. Biochemical test results showed that the levels of total testosterone were significantly higher in the group of patients with PCOS. It was found that FSH levels were significantly lower, and the levels of LH and anti-Mullerian hormone (AMH) were significantly higher in women with PCOS. In addition, the patients with PCOS showed significantly increased levels of 17-OH-progesterone and BMI compared with the control group. The levels of sex hormone binding globulin (SHBG) were significantly lower in women with PCOS.
Despite the fact that these important data confirm involvement of the LHB gene in the pathogenesis of PCOS, the study results in different ethnic groups and in different genomic regions demonstrate heterogeneity. It was found that changes in the LHB gene can influence the structure and function of LH, thereby changing its biological activity and leading to the disorders, such as anovulation, the absence of menstruation, and polycystic ovaries in women [4]. It is important to note that the luteinizing hormone beta-subunit exon 3 (Gly102Ser) gene mutation is rare in Korean women with endometriosis and polycystic ovary syndrome [13].
Our study analyzed frequency distribution of alleles and genotypes in replacement of Gly102Ser (rs5030774) in the LHB gene in women with PCOS and in the control group.
Women in both groups had the G/G genotype (Table 3).

In the course of our research, the rs5030774 polymorphic locus of the LHB gene was not identified and did not show the association with the development of PCOS. It is known that the Gly102Ser missense mutation in the LHB gene can reduce LH activity, that contradicts the pathogenesis of PSCOS, which is characterized by elevated level of LH versus FSH.
According to the 1000 Genomes Project, the proportion of the minor A allele of this variant in the LHB gene is extremely small – only 0.01% [14].
Further, we analyzed allele frequency and genotype distribution of the polymorphic loci rs5030646 and rs11031006 of the FSHB gene in patients with PCOS and in the control group (Table 3).
The rs5030646 polymorphic locus of the FSHB gene was detected neither in the group of women with PCOS, not in the control group. According to the 1000 Genomes Project, the minor G allele frequency of the FSHB gene (rs5030646) is also extremely low – less than 0.01% [14].
The rs5030646 variant in the FSHB gene represents a shift in reading frame, that leads to formation of proteins with shorter length. This gene variant is classified as "pathogenic" and is associated with reduced FSH level in the blood. Given that FSH deficiency prevents normal follicle maturation, it can be assumed that the defective protein formed as a result of this gene variant can be a cause of infertility. A case of infertility associated with similar deletion in the FSHB gene, as well as similar clinical manifestations with a heterozygous genotype have been described in scientific literature [15].
Our study showed that allele and genotype distributions of the rs11031006 variant in the FSHB gene in the analyzed samples complied with the Hardy–Weinberg equilibrium (Table 3). Detailed analysis of the genotype and allele frequencies found statistically significant differences between the groups under study (p=0.02 and p=0.008, respectively) (Table 3).
The A allele was more common (59/110 (53.6%)) in the control group compared with the group of women with PCOS (41/114 (36.0%)). Higher frequencies of the A allele and the homozygous AA genotype in the control group suggest that their carriers are at low risk of developing PCOS; and the A allele and AA genotype suggest their protective role. Given that PCOS is characterized by the imbalanced ratio between LH and FSH, it is possible that the A allele of the FSHB gene contributes to maintaining hormonal balance by increasing gene activity and preventing reduction in FSH level, that has been recently confirmed by Bohaczuk S.C. et al. [16].
The rs11031006 variant is located in the promoter region of the FSHB gene and is characterized by replacement of the guanine base with adenine at position 211. One of meta-analyses found a significant association between this variant in the FSHB gene and PCOS due to elevated LH level [17]. However, another subsequent study did not confirm this relationship. The authors found an increase, but not reduction, in FSHB activity in carriers of the minor A allele [16].
The frequency of the A allele in the studied samples of women – 59/110 (53.6%) in the control group and 41/114 (36.0%) in the group with PCOS, significantly exceeded the allele frequency in the worldwide population registered in the 1000 Genomes Project, and was 0.08% [14]. The highest occurrence of this allele is observed in the Finnish population (18%). At the same time, it is much less common in the East Asian population (3%).
Distribution of genotypes and alleles of the rs4953616 polymorphic locus of the LHCGR gene in the control groups and in women with PCOS complied with the Hardy–Weinberg Law of Genetic Equilibrium (Table 3). The homozygous T/T genotype was significantly more common in the group of women with PCOS – 36/57 (63.2%) versus 20/55 (36.4%) in the control group. On the contrary, the heterozygous C/T genotype prevailed among healthy women – 23/55 (41.8%) (Table 3).
The data obtained by us indicate association between the T allele and PCOS (86/110 (76.3%) at p=0.002; OR=2.404, 95% CI 1.354–4.267). At the same time, an increased frequency of the TT genotype was observed in women with PCOS (72/114 (63.2%) at p=0.016; OR=3.000, 95% CI 1.390–6.473). This indicate that the T allele and TT genotype can be attributed to risk factors for PCOS.
The LHCGR receptor acts as a high affinity-receptor for LH and promotes steroidogenesis and follicle maturation [18]. The rs4953616 variant located in intron 6 of the LHCGR gene can influence this gene expression or change the properties of the encoded protein. The T allele in the LHCGR gene can potentially change intracellular signaling after exposure to LH. This, in turn, can lead to dysregulation of follicle growth, development and viability, that is of great importance for normal function of the ovaries. These changes can be associated with cyst formation, which is characteristic of PCOS. The T allele has been shown to be associated with the risk of developing PCOS [18]. In particular, the study conducted with a sample of Arab women found statistically significant association between the T allele of the rs4953616 variant in the LHCGR gene and PCOS. At the same time, the proportion of both TT homozygotes and CT heterozygotes was higher compared with the control group [19].
According to the 1000 Genomes Project, the frequency of occurrence of the C allele of the rs4953616 variant in the LHCGR gene in the general population is 32% [14]. At the same time, the T allele is most common in European (75%) and East Asian (77%) populations. Such a significant variability in the prevalence of this variant in the LHCGR gene in different ethnic and geographical groups indicates the need to take into account the population specificity in exploring its association with PCOS.
The genotype distribution of the rs6165 variant in the FSHR gene in women with PCOS and in the control group complied with the Hardy–Weinberg law (Table 3). Statistical analysis showed significant differences in genotype frequencies between the control group and the group of women with PCOS (p=0.036), as well as in distribution of rs6165 alleles in the FSHR gene (p=0.014) (Table 3). In the control group, the T allele was more common – 86/110 (78.2%) versus 72/114 (63.2%) in the group with PCOS, thus suggesting their protective role.
The rs6165 variant in the FSHR gene is a missense variant located in exon 10, that leads to substitution of alanine for threonine at the 307th amino acid position. These amino acids have different biochemical characteristics. Threonine contains a hydroxyl group, that can form hydrogen bonds and, thereby influence protein structure, its interaction with other molecules, including FSH ligands. Therefore, it is most likely that this missense variant affects receptor function [20].
PCOS is often associated with imbalance in LH and FSH, and reduced sensitivity to FSH can increase the likelihood of this imbalance, especially when the levels of these hormones in the blood are stable [21].
The study on the Chinese population did not confirm the association between the rs6165 polymorphic locus of the FSHR gene and PCOS. Nevertheless, the researchers noted significantly different distribution of two-marker haplotypes combining the rs6165 and rs6166 variants (p=0.042) versus the control group. This indicates that polymorphic loci acting in a close relationship can be associated with the disease [22]. This effect suggests that a single genetic change does not have a noticeable effect, but combined genetic mutations can modify biological processes and mechanisms, that ultimately contribute to the development of PCOS. The association between the rs6165 polymorphic locus of the FSHR gene and PCOS was found in women in our study.
According to the 1000 Genomes Project, the percentage of cases with rs6165 variant in the FSHR gene is approximately 50% worldwide. At the same time, the T allele is most common among the East Asian population (66%) [14]. The association between the rs6165 polymorphic locus of the FSHR gene and fertility problems, including PCOS, can depend on the ethnicity. Due to this, it is important to take into account the population specificity when conducting research.
Conclusion
Exploration of polymorphic loci of gonadotropin genes and their receptor gene variants showed statistically significant association between PCOS and rs11031006 in the FSHB gene, rs4953616 in the LHCGR gene, and rs6165 in the FSHR gene.
References
- Беглова А.Ю., Елгина С.И., Гордеева Л.А. Полиморфизм генов CYP11A1, CYP17, CYP19 у женщин репродуктивного возраста с синдромом поликистозных яичников. Акушерство и гинекология. 2019; 12: 148-53. [Beglova A.Yu., Elgina S.I., Gordeeva L.A. Polymorphism of the CYP11A1, CYP17A1, and CYP19A1 genes in reproductive-aged women with polycystic ovary syndrome. Obstetrics and Gynecology. 2019; (12): 148-53 (in Russian)]. https://dx.doi.org/10.18565/aig.2019.12.148-153
- Найдукова А.А., Каприна Е.К., Донников А.Е., Чернуха Г.Е. Генетические аспекты формирования синдрома поликистозных яичников. Акушерство и гинекология. 2016; 3: 16-22. [Naidukova A.A., Kaprina E.K., Donnikov A.E., Chernukha G.E. Development of polycystic ovary syndrome: Genetic aspects. Obstetrics and Gynecology. 2016; (3): 16-22 (in Russian)]. https://dx.doi.org/10.18565/aig.2016.3.16-22
- GWAS Catalog. Trait: polycystic ovary syndrome. Available at: https://www.ebi.ac.uk/gwas/efotraits/EFO_0000660
- Koloda Y.A., Denisova Y.V., Podzolkova N.M. Genetic polymorphisms of reproductive hormones and their receptors in assisted reproduction technology for patients with polycystic ovary syndrome. Drug Metab. Pers. Ther. 2021; 37(2): 111-22. https://dx.doi.org/10.1515/dmpt-2021-0123
- Bashir K., Anum A., Idrees I., Manzoor H.T. Elucidating the molecular genetics of genes CYP19A1, CYP17, and FSHR variants association in polycystic ovarian syndrome. Rus. J. Genet. 2024; 60(3): 387-97. https://dx.doi.org/10.1134/S1022795424030049
- Chen Z.J., Zhao H., He L., Shi Y., Qin Y., Shi Y. et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16. 3, 2p21 and 9q33. 3. Nat. Genet. 2011; 43(1): 55-9. https://dx.doi.org/10.1038/ng.732
- Deswal R., Nanda S., Dang A.S. Association of luteinizing hormone and LH receptor gene polymorphism with susceptibility of polycystic ovary syndrome. Syst. Biol. Reprod. Med. 2019; 65(5): 400-8. https://dx.doi.org/10.1080/19396368.2019.1595217
- Dou Y., Zhao R., Wu H., Yu Z., Yin C., Yang J. et al. DENND1A desensitizes granulosa cells to FSH by arresting intracellular FSHR transportation. Sci. China Life Sci. 2024; 67(8): 1620-34. http://dx.doi.org/10.1007/s11427-023-2438-4
- Shi Y., Zhao H., Shi Y., Cao Y., Yang D., Li Z. et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat. Genet. 2012; 44(9): 1020-5. http://dx.doi.org/10.1038/ng.2384
- Smet M.E., McLennan A. Rotterdam criteria, the end. Australas. J. Ultrasound Med. 2018; 21(2): 59-60. https://dx.doi.org/10.1002/ajum.12096
- Гарафутдинов Р.Р., Баймиев Ан.Х., Малеев Г.В., Алексеев Я.И., Зубов В.В., Чемерис Д.А., Кирьянова О.Ю., Губайдуллин И.М., Матниязов Р.Т., Сахабутдинова А.Р., Никоноров Ю.М., Кулуев Б.Р., Баймиев Ал.Х., Чемерис А.В. Разнообразие праймеров для ПЦР и принципы их подбора. Биомика. 2019; 11(1): 23-70. [Garafutdinov R.R., Baymiev An.Kh., Maleev G.V., Alexeyev Ya.I., Zubov V.V., Chemeris D.A., Kiryanova J.Yu., Gubaydullin I.M., Matniyazov R.T., Sakhabutdinova A.R., Nikonorov Yu.M., Kuluev B.R., Baymiev Al.Kh., Chemeris A.V. Diversity of PCR primers and principles of their design. Biomics. 2019; 11(1): 23-70 (in Russian)]. https://dx.doi.org/10.31301/2221-6197.bmcs.2019-04
- Petrie A., Bulman J.S., Osborn J.F. Further statistics in dentistry. Part 8: Systematic reviews and meta-analyses. Br. Dent. J. 2003; 194(2): 73-8. https://dx.doi.org/10.1038/sj.bdj.4809877
- Kim N.K., Lee E.G., Cho M.S., Nam Y.S., Chung H.M., Chung K.W. et al. Analysis of LHbeta exon 3 (Gly102Ser) gene mutation in infertile patients with endometriosis and polycystic ovary syndrome (PCOS). Kor. J. Fertil. Steril. 2001; 3(27): 291-4.
- Ensembl genome browser. Available at: https://www.ensembl.org/index.html
- Liaqat I., Jahan N., Krikun G., Taylor H.S. Genetic polymorphisms in Pakistani women with polycystic ovary syndrome. Reprod. Sci. 2015; 22(3): 347-57. http://dx.doi.org/10.1177/1933719114542015
- Bohaczuk S.C., Thackray V.G., Shen J., Skowronska-Krawczyk D., Mellon P.L. FSHB transcription is regulated by a novel 5′ distal enhancer with a fertility-associated single nucleotide polymorphism. Endocrinology. 2021; 162(1): bqaa181. https://dx.doi.org/10.1210/endocr/bqaa181
- Hayes M.G., Urbanek M., Ehrmann D.A., Armstrong L.L., Lee J.Y., Sisk R. et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat. Commun. 2015; 6: 7502. https://dx.doi.org/10.1038/ncomms8502
- Singh S., Kaur M., Beri A., Kaur A. Significance of LHCGR polymorphisms in polycystic ovary syndrome: an association study. Sci. Rep. 2023; 13(1): 22841. http://dx.doi.org/10.1038/s41598-023-48881-0
- Almawi W.Y., Hubail B., Arekat D.Z., Al-Farsi S.M., Al-Kindi S.K., Arekat M.R. et al. Leutinizing hormone / choriogonadotropin receptor and follicle stimulating hormone receptor gene variants in polycystic ovary syndrome. J. Assist. Reprod. Genet. 2015; 32(4): 607-14. https://dx.doi.org/10.1007/s10815-015-0427-0
- Renzi A., Petersen C.G., Vagnini L., Oliveira-Pelegrin G.R., Mauri A.L., Massaro F.C. et al. The fshr gene polymorphism (rs 6165-ala/ala genotype) is associated with the use of higher doses of recombinant FSH during IVF/ICSI treatment. Fertil. Steril. 2014; 102(3): e120-1. https://dx.doi.org/10.1016/j.fertnstert.2014.07.412
- Kim J.J., Choi Y.M., Hong M.A., Chae S.J., Hwang K., Yoon S.H. et al. FSH receptor gene p. Thr307Ala and p. Asn680Ser polymorphisms are associated with the risk of polycystic ovary syndrome. J. Assist. Reprod. Genet. 2017; 34(8): 1087-93. https://dx.doi.org/10.1007/s10815-017-0953-z
- Du J., Zhang W., Guo L., Zhang Z., Shi H., Wang J. et al. Two FSHR variants, haplotypes and meta-analysis in Chinese women with premature ovarian failure and polycystic ovary syndrome. Mol. Genet. Metab. 2010; 100(3): 292-5. https://dx.doi.org/10.1016/j.ymgme.2010.03.018
Received 10.05.2025
Accepted 30.07.2025
About the Authors
Ksenia A. Ustenko, student, Rostov State Medical University, Ministry of Health of Russia, 344022, Russia, Rostov-on-Don, per. Nakhichevansky, 29, +7(928) 770-33-34, ustenko.ksusha@gmail.com, https://orcid.org/0009-0003-3992-1923Ekaterina G. Derevyanchuk, PhD, Associate Professor, Southern Federal University, 344090, Russia, Rostov-on-Don, Stachki Ave., 194 build. 1, +7(928)132-29-88, biolab2008@yandex.ru, https://orcid.org/0000-0002-6231-3454
Anzhela A. Aleksandrova, PhD, Associate Professor, Southern Federal University, 344090, Russia, Rostov-on-Don, Stachki Ave., 194 build. 1, +7(928)190-58-30, aalexsandrova@mail.ru, https://orcid.org/0000-0002-1948-4995
Corresponding author: Ekaterina G. Derevyanchuk, biolab2008@yandex.ru



