Development and validation of models to predict total and early-onset preeclampsia in the first trimester of pregnancy using machine learning algorithms
Andreychenko A.E., Luchinin A.S., Ivshin A.A., Ermak A.D., Novitskiy R.E., Gusev A.V.
Background: Preeclampsia (PE) is a life-threatening and difficult-to-predict complication of pregnancy, characterized by multi-organ dysfunction. PE affects 2–8% of all pregnancies and is one of the leading causes of perinatal and maternal mortality, especially in cases of early onset PE. Objective: To develop models for predicting total and early onset PE in the first trimester of pregnancy using machine learning (ML) technologies based on real-world clinical data. Materials and methods: We analyzed 21,092 records obtained from electronic medical records through the Webiomed platform, corresponding to 12,434 unique pregnancies of 12,283 women aged 11 to 60 years, up to 16 weeks. Anamnestic, constitutional, clinical, instrumental, and laboratory data, commonly used in routine medical practice, were selected as potential factors for predicting PE, totaling 53 variables. To create the models, we employed logistic regression (LR), gradient boosting methods (LightGBM, XGBoost, CatBoost), and methods based on decision trees (RandomForest and ExtraTrees). Results: The ExtraTrees model demonstrated the highest accuracy in predicting PE, with an area under the curve (AUC) of 0.858 (95% CI 0.827–0.890). The model's overall accuracy was 0.634 (95% CI 0.616–0.652), sensitivity was 0.897 (95% CI 0.837–0.953), and specificity was 0.624 (95% CI 0.605–0.643). Among the models for assessing the risk of early onset PE, the RandomForest algorithm yielded the most promising results. The AUC after validation was 0.848 (95% CI 0.785–0.904), with an accuracy of 0.813 (95% CI 0.798–0.828), sensitivity of 0.733 (95% CI 0.565–0.885), and specificity of 0.814 (95% CI 0.799–0.828). Conclusion: The metrics of the final models align with previously published models. External validation results demonstrate the relative stability of the models with new data, indicating their potential applicability in real clinical practice. This is our first experience in predicting complex pregnancy complications based on real-world clinical data. The quality of the predictive model depends directly on the data and the statistical algorithms used, aspects that we intend to refine in future studies.
Authors' contributions: Andreychenko A.E. – study administration, manuscript editing; Luchinin A.S. – manuscript drafting; Ivshin A.A. – conception of the study, expert analysis of results, manuscript editing; Ermak A.D. – modeling and data analysis; Novitskiy R.E., Gusev A.V. – conception of the study.
Conflicts of interest: The authors have no conflicts of interest to declare.
Funding: This study was performed using the Unique Scientific Unit (UNU) «Multicomponent software and hardware system for automated collection, storage, markup of research and clinical biomedical data, their unification and analysis based on Data Center with Artificial Intelligence technologies» (reg. number: 2075518).
Acknowledgments: The authors would like to thank D.V. Gavrilov for valuable consultations during data collection and validation of model results, and to V.O. Barkina and V.Yu. Borisov for assistance in data processing and model building.
Authors' Data Sharing Statement: The data supporting the findings of this study are available upon request from the corresponding author after approval from the principal investigator.
For citation: Andreychenko A.E., Luchinin A.S., Ivshin A.A., Ermak A.D., Novitskiy R.E., Gusev A.V. Development and validation of models to predict total and early-onset preeclampsia in the first trimester of pregnancy using machine learning algorithms. Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2023; (10): 94-107 (in Russian) https://dx.doi.org/10.18565/aig.2023.101
Keywords
Preeclampsia (PE) is a life-threatening and difficult-to-predict complication of pregnancy that is characterized by multi-organ dysfunction. It affects 2–8% of all pregnancies and is a leading cause of perinatal and maternal mortality [1]. Currently, the etiology and pathogenesis of PE are not fully understood and therapy remains symptomatic. PE progresses differently in various patients because different organ systems are involved in the pathological process. However, the clinical presentation of multiple organ dysfunction varies widely [2]. This diversity makes timely diagnosis and prediction of the onset and progression of PE challenging. In this context, the creation and implementation of a multifactorial model for predicting PE in real-world clinical practice is urgently needed to enhance pregnancy outcomes [3]. A recent systematic review of 68 PE predictive models revealed that the most commonly used predictors were pregnancy history, body mass index (BMI), blood pressure (BP), uterine artery flow status, and maternal age. Conversely, specific biomarkers and predictors from the ultrasound diagnostic results did not significantly improve the discriminative properties of the models. The area under the ROC curve (AUC) for the studied models varied widely, ranging from 0.61 to 0.996. Only 4% and 6% of all papers included information on internal and external validation, respectively [4].
Despite the existing difficulties, the construction of multiparametric prognostic models can contribute to the effective prediction of PE, ultimately enabling timely clinical decision-making for effective prevention. Notably, studies related to the development of predictive models for PE have been published in scientific literature. For instance, Thangaratinam et al. constructed two predictive models (PREP-L and PREP-S) using Cox regression analysis and logistic regression, incorporating parameters associated with the impact of treatment for high blood pressure and antispasmodics. However, owing to their complexity and inconvenience, these models have not yet been integrated into clinical practice [5]. One option to simplify, make more convenient, and intuitively understand prognostic models is their graphical representation as a nomogram, a method frequently employed in oncology [6]. This approach, combined with the LR algorithm, formed the basis of another PE prediction model with commendable discriminative ability. Internal validation yielded an AUC of 0.957 (0.935–0.979) and a mean absolute error of 1.4% for the calibration curve [7].
Given that the incidence of PE persists worldwide, the quality and effectiveness of the developed prediction models vary widely, and practical applications remain limited. Therefore, continued research in this area remains relevant.
This study aimed to develop models for predicting both total and early onset PE in the first trimester of pregnancy using machine learning (ML) technologies based on real-world clinical data.
Materials and methods
Data Source. This retrospective study was conducted using the Webiomed predictive analytics platform database, which contains non-personalized formalized data from electronic medical records (EMRs) of 11.6 million patients who underwent examinations and treatments in various medical organizations across different regions of the Russian Federation. To collect the data, the Webiomed platform developer signed agreements with relevant operators of personal medical data to anonymize them on the operator's side and transfer the results to the Webiomed platform, including for research purposes. Since anonymized medical data were analyzed, informed voluntary consent from the patients was not required. Medical data and machine-readable variable values were extracted from EMRs using various technologies, including Natural Language Processing (NLP). At the time of the study, the Webiomed platform supported the automatic extraction of over 2,900 logical, categorical, and quantitative variables.
Participants. A dataset of 21,092 records corresponding to 12,434 unique pregnancies of 12,283 women who received medical care between 12/21/2004 and 11/28/2022 was generated from the existing database. A record was defined as a case of medical care recorded in the patient's EMR with the gestational age established at the time of referral, together with the values of the patient's clinical parameters updated as of the date of the case. The following criteria were used to include a case in the dataset: 1) established diagnosis corresponding to the pregnancy status according to ICD-10 classification (refer to Table 1); 2) gestational age in the range of 7–16 weeks; 3) known pregnancy outcome (based on EMR with appropriate ICD-10 codes); and 4) mandatory presence of values for age, systolic BP, diastolic BP, height, and weight.
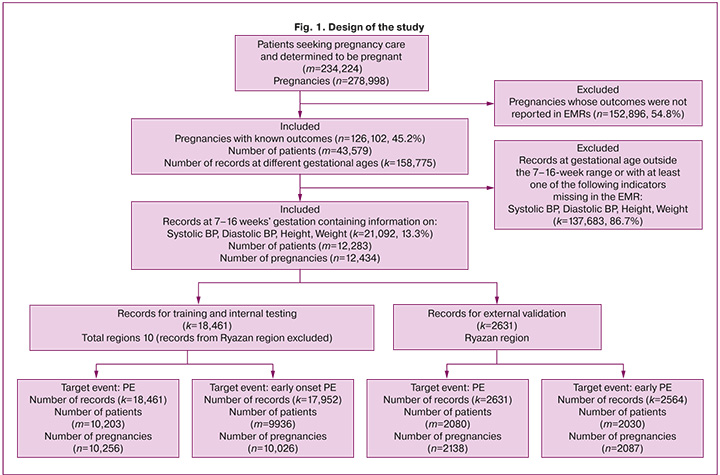
From the final dataset, a separate sample was generated to include records pertaining only to pregnancies with early onset PE or no PE (total of 20,516 medical records). The resulting datasets contained information on pregnancies in patients from 11 regions of the Russian Federation. Based on the territorial distribution for both groups, samples were excluded for external validation [8], which included records of pregnant women from the Ryazan region: 2,564 records for predicting early onset PE and 2,631 records for determining the overall risk of PE during pregnancy. The sample for external validation was formed according to the Type 2b design of the TRIPOD statement [8]. According to these guidelines, sampling for external validation based on area or time period is a stronger design for the external validation of predictive models than randomization. The overall design of this study is illustrated in Figure 1. After excluding the Ryazan region data, the remaining data were used for model development and randomly divided into a training sample (80%), a sample for hyperparameter selection (10%), and internal testing (10%). Samples for internal testing and external validation were not used to train the algorithms.
In the current study, PE prediction models were created based on multicenter data from real clinical practice in different regions of the Russian Federation, without imposing a requirement for the mandatory availability of the results of individual laboratory tests that have known prognostic value with respect to PE prediction but are very rarely used in daily practice.
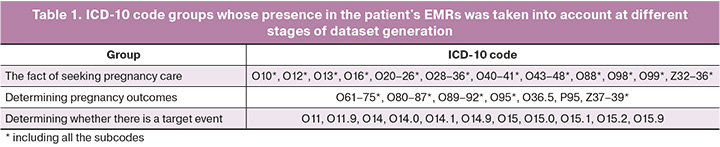
Outcomes. PE was the target event in this study. All patient records pertaining to the current pregnancy were assigned to the group with the target event if PE was diagnosed during the course of pregnancy. If no ICD-10 codes selected for the target event were present in the patient's EMR during the current pregnancy, all records corresponding to the pregnancy were assigned to the group without the target event. The ICD-10 codes used to select patients who sought care for pregnancy, determine pregnancy outcomes, and determine the presence or absence of the target event are summarized in Table 1.
Predictors. Anamnestic, constitutional, clinical, and laboratory parameters (53 variables in total) were selected as potential predictors of PE. The dataset included parameter values from the EMRs of patients enrolled no later than the 16th week of gestation. Considering that PE is not diagnosed before the 20th week of gestation, parameters studied between the 7th and 16th weeks of pregnancy were considered prognostic factors for PE. Anamnestic factors include concomitant or past diseases, pathologic conditions, menstrual cycle patterns, parity, and habits that could affect the outcome of the current pregnancy, including tobacco smoking, age at menarche, upcoming first birth, infertility, in vitro fertilization (IVF), low birth weight, PE, placental insufficiency, fetal growth retardation, history of fetal distress and death, intergenic interval, stillbirth, cardiovascular disease (CVD), thrombosis, neurologic disease, urinary tract disease (UTD), diabetes mellitus (DM), gestational DM, and sexually transmitted infections (STIs). Constitutional parameters included age at the onset of pregnancy, height, weight during and before pregnancy, BMI during and before pregnancy, and gestational age at the time of analysis. Clinical factors that characterized the current pregnancy included skin cyanosis, visual disturbance, headache, abdominal pain, cramps, nausea, vomiting, systolic BP (SBP), diastolic BP (DBP), mean BP, severe arterial hypertension (AH), moderate AH, edema, pulmonary edema, hemolysis, anuria or oliguria, multiorgan failure, anemia, thrombocytopenia, and multiple births. Laboratory parameters included platelet count, blood creatinine, alanine aminotransferase (ALT), aspartate aminotransferase (AST), activated partial thromboplastin time (APTT), and fibrinogen.
Correction of Outliers and Filling in Missing Values. Filling of missing values in numerical parameters was performed using the constant value "-10000" [9] and in binary parameters, with zero values (indicating the absence of the outcome). When processing the quantitative variables, values beyond the limits established in clinical practice were excluded. The parameter value bounds used to remove outliers are presented in Supplementary tables. In addition, the numerical parameters were transformed using histogram normalization before modeling [10].
Statistical analysis
Statistical analysis and ML model building were conducted using Python programming language, version 3.9. The normality of the distribution of quantitative variables was assessed using the Shapiro–Wilk test; their data are presented as medians with interquartile ranges, while categorical variables are shown as proportions (N, %). Comparison of quantitative variables in groups with and without the target event (PE) was performed using the Mann–Whitney test, and categorical variables were compared using χ²; a p-value < 0.05 was considered statistically significant.
Logistic regression (LR), gradient boosting methods (LightGBM, XGBoost, CatBoost), and methods based on decision trees (Random Forest and ExtraTrees) were employed as ML algorithms. Internal validation of the obtained models was performed using a test dataset. AUC, sensitivity (Recall), specificity, accuracy (Accuracy), positive predictive value (Precision), F-measure, error matrix, and calibration curves served as criteria for assessing the efficiency and quality of the models [11, 12]. The confidence intervals of the selected statistics were estimated using the bootstrap method by randomly generating a set of pseudo-samples [13]. The maximum Youden index was used as the threshold for classifying the results of the model performance. The significance of the predictors included in the ML models was determined using the SHAP method [14].
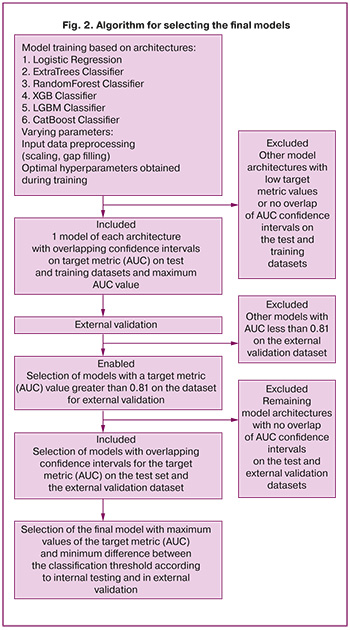
The models selected according to our algorithm were subjected to external validation. The calculation of the error matrix and all quality metrics in this case was performed using the classification threshold according to internal testing and the maximum Youden index determined on the dataset for external validation. The selection of the final model was based on the maximum AUC value in the external validation, intersection of the metric confidence intervals, and minimum difference between the classification thresholds according to internal testing and external validation. The complete model-selection procedure is illustrated in Figure 2.
Results
Descriptive statistics. Following the generation of the dataset for training and internal testing (i.e., development), 672 (3.6%) records pertaining to 358 pregnancies during which PE was diagnosed (class 1) were included in the group with the presence of the target event. The remaining 17,789 (96.4%) records (9,938 pregnancies) were assigned to the group without a target event (Grade 0). When forming the subsample for predicting the risk of early onset PE, records pertaining to pregnancies in which PE developed after week 34 were excluded from the total number of records with a target event. The total number of records of a target event in this group was 163 (0.9%).
The distribution in terms of frequency of occurrence and magnitude of a number of variables in the total dataset between the two classes showed statistically significant differences. The classes differed in terms of characteristics such as weight and BMI before and during pregnancy, levels of SBP and DBP, and mean BP and ALT. In addition, among the records with PE (class 1), indications of headache, nausea, vomiting, abdominal pain, edema, hemolysis, visual disturbances, moderate-to-severe AH, low birth weight, fetal growth retardation and distress, placental insufficiency, CVD, neurological diseases, and DM were more common. Among the patients who formed class 1 records, there were more primiparous women and women with an aggravated history of PE and infertility. The statistical measures of the magnitude and frequency of the variables in the training and testing datasets according to the presence of the target event are presented in Table 2.
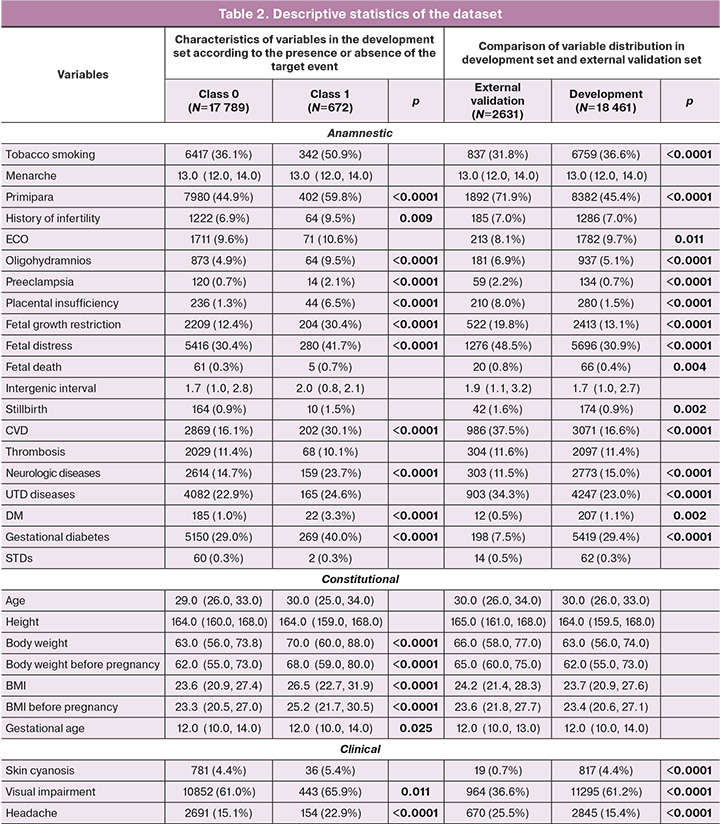
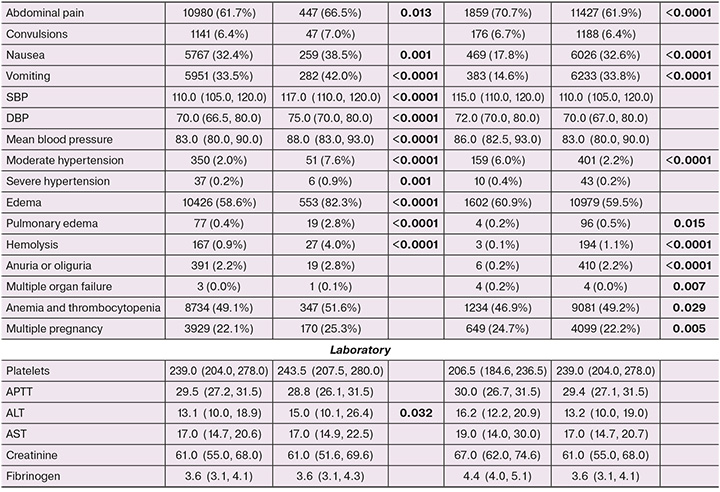
The external validation dataset included 2631 patient records from the Ryazan region. A class division of records corresponding to the development set was observed: class 1 – 97 (3.6%); class 0 – 2534 (96.3%). The development and external validation sets of early onset PE prediction models also had the same distribution by disease development; the proportion of records with a target event was 1% (163 and 30 records, respectively).
When comparing the distribution and frequency of occurrence of the selected predictors between the two sets, statistically significant differences were found in the frequency of UTD, DM, STIs, neurological diseases, CVD, placental insufficiency, fetal distress, low birth weight, tobacco smoking, and a number of factors characterizing the course of the current pregnancy (abdominal pain, headache, nausea, visual disturbance, multiorgan failure, skin cyanosis, hemolysis, and multiple pregnancies). The general characteristics of the datasets used and their comparisons are presented in Table 2.
Model development. Model development for predicting PE and early onset PE during pregnancy was performed using six ML algorithms: LR, LightGBM, XGBoost, CatBoost, Random Forest, and ExtraTrees. Based on the absolute values of the Shepley vectors obtained by training the algorithms on the initial datasets, variable selection was performed, selecting predictors that contributed 95% of the cumulative percentages to the models' results for further analysis. In addition, irrespective of SHAP significance, a list of obligatory variables determined based on generally accepted maternal risk factors for PE was retained for the study [15].
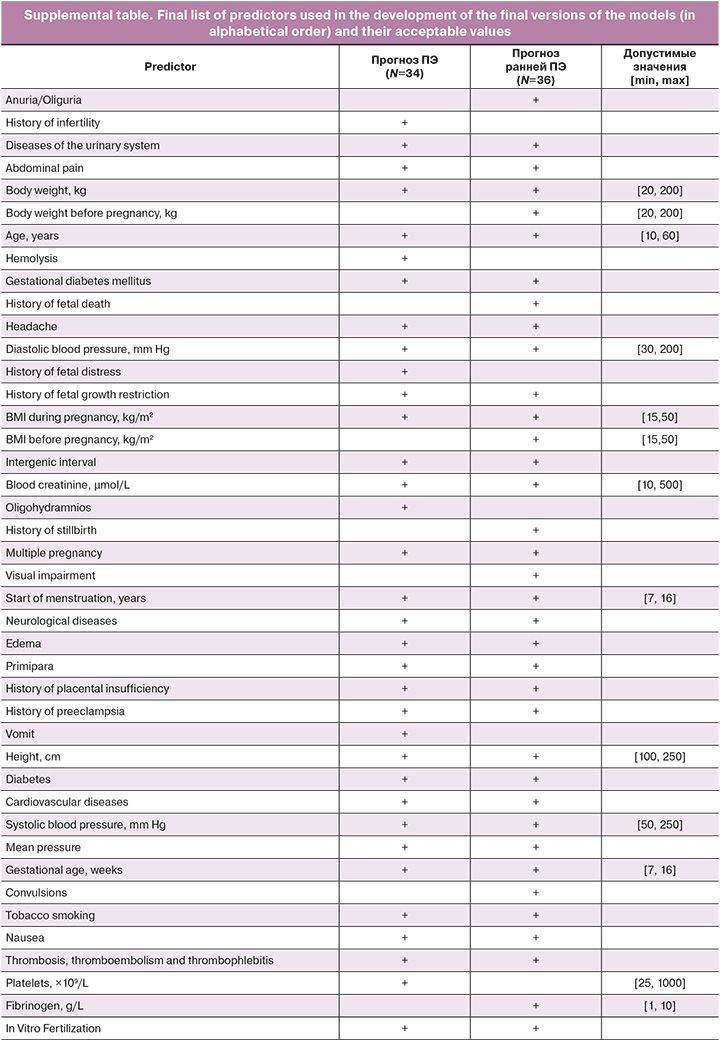
The next step was to retrain the algorithms by considering only selected variables. For the PE prediction models, 34 predictors were included in the final list, whereas 36 predictors were included for early onset PE. The list of variables used as predictors in the models is presented in the Appendix. Only models that showed the maximum value of the target quality metric (AUC) on the test dataset at the intersection of the 95% confidence intervals of this metric on the training and test samples were retained for further study to avoid overtraining. Based on the external validation results, the final models were selected according to the algorithm shown in Figure 2.
Model Performance. The characterization of the performance of each model after applying the classification threshold to the internal test set and external validation are presented in Tables 3 and 4, respectively. The ExtraTrees model showed the highest discriminatory ability and robustness to external data in the task of predicting up to and including 16 weeks' gestational PE development during pregnancy, with a target metric AUC value of 0.858 (95% CI 0.827–0.890) for external validation and 0.862 (95% CI 0.800–0.914). For the external dataset, the accuracy of this model with a classification threshold of 0.04 was 0.634 (95% CI 0.616–0.652), sensitivity was 0.897 (95% CI 0.837–0.953), and specificity was 0.624 (95% CI 0.606–0.643).
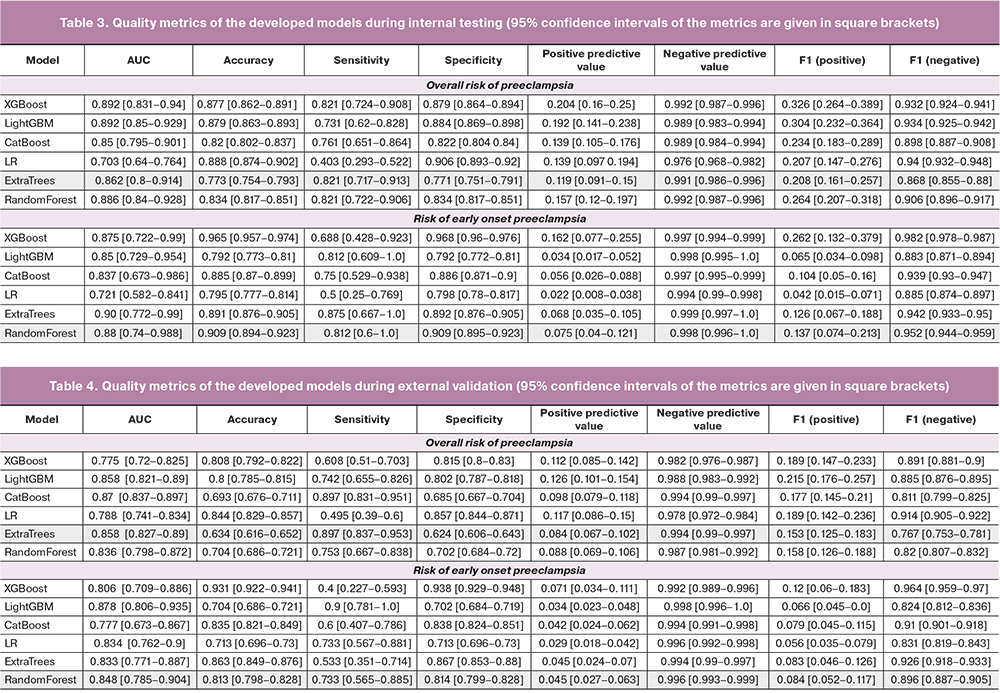
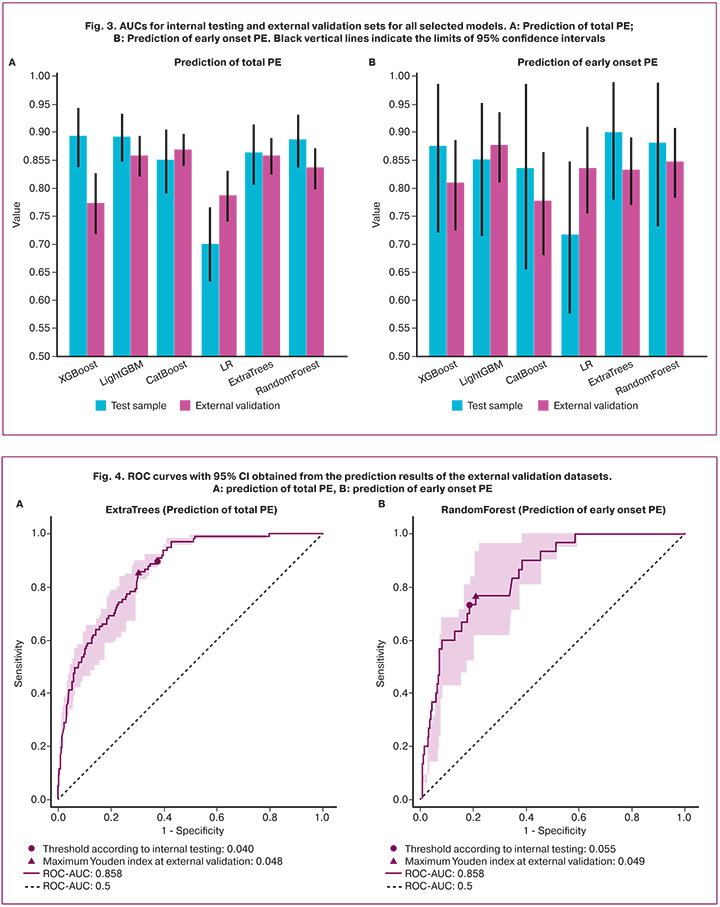
Among the models for predicting the development of early onset PE, the model based on the random forest algorithm performed the best. The following quality metrics were obtained by external validation at a binarization threshold of 0.055: AUC 0.848 (95% CI 0.785–0.904), accuracy 0.813 (95% CI 0.798–0.828), sensitivity 0.733 (95% CI 0.565–0.885), specificity 0.814 (95% CI 0.799–0.828). The AUC values obtained for the external and internal validation sets for all selected models are presented in Figure 3. The ROC curves of the selected final models are shown in Figure 4.
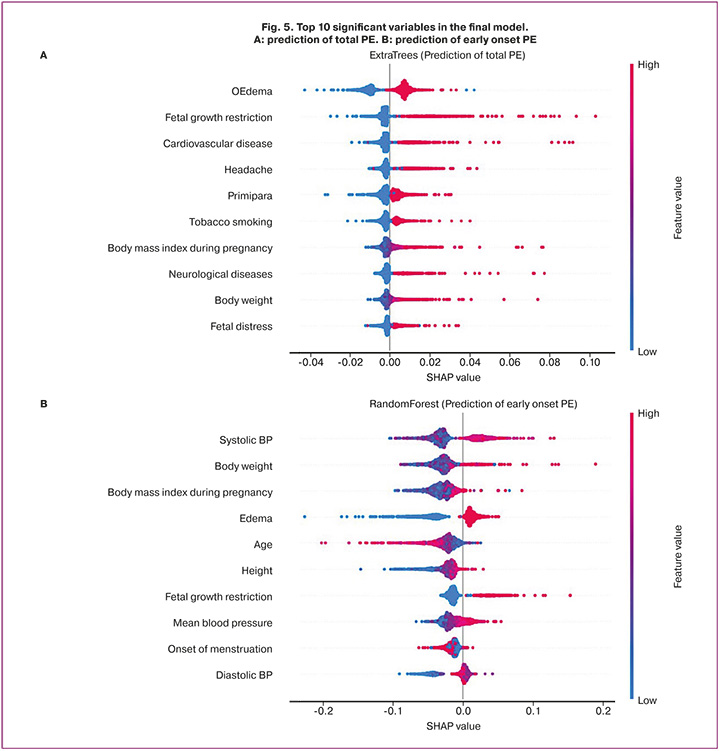
The significance of the 10 key variables included in the selected ExtraTrees and Random Forest models is shown in Figure 5. The mean absolute errors of the calibration curve for the selected final PE and early onset PE prediction models were 24.5% and 22.4%, respectively.
Thus, the developed ML models can automatically analyze the above signs from the patients' EMRs, provided that the gestational age is between 7–16 weeks, and detect a high risk of PE if the results of the analysis exceed the set activation thresholds. The provided red flag system is able to alert the physician of a high risk of PE and the need to consider prophylaxis.
Discussion
PE is a serious complication of pregnancy that poses a threat to the health and lives of both mother and child. However, the pathogenesis of PE remains unclear. Various factors, including maternal, fetal, and placental, play a role in the development of this pregnancy complication. However, none of the studies [7, 16] can fully elucidate the pathogenesis of PE or reliably predict its occurrence.
During PE, placental trophoblastic invasion is disrupted, resulting in reduced oxygen supply to the placenta. This leads to the release of inflammatory factors from the vascular endothelium and the subsequent damage. These events trigger systemic spasms of the small vessels and diminished perfusion in organs, ultimately leading to organ dysfunction. This cascade of events can culminate in the development of conditions such as eclampsia, placental abruption, and, in severe cases, mortality of both the mother and fetus [2, 17].
The concept of early onset PE occurring before the 34th week of pregnancy warrants special attention because of its significant clinical and pathogenetic distinctions from late-onset PE. Early onset PE is a major contributor to perinatal mortality [15]. Thus, early prediction of both total and early onset PE, as well as timely medical intervention and prevention, are of paramount importance in reducing the incidence of this complication and improving pregnancy outcomes.
Several multiparameter machine learning (ML) models have been developed, and their accuracy has been tested. Among these models, those based on decision trees demonstrated the highest performance in terms of the target quality metric using the collected data. The detection of edema, headaches during pregnancy, and monitoring of blood pressure levels are crucial factors for these models. Notably, constitutional and anthropometric data of the patients, including age, weight, height, and BMI during pregnancy, were significant features. Additionally, anamnestic data, such as cardiovascular diseases, neurological conditions, fetal growth restriction, and age at menarche, played a pivotal role in the model.
The positive class predictive value of the resulting model for assessing the risk of total PE in the internal validation ranged from 9% to 15%, and for predicting early onset PE, it ranged from 4% to 11%. However, according to the results of the external validation, a decrease in these indicators was observed, specifically from 6% to 10% and 3% to 7%, respectively. Nevertheless, the tools we developed demonstrated high sensitivity and negative predictive value (both exceeding 99% for both models, according to the results of external validation). This high accuracy is crucial for ruling out such complications during pregnancy in the diagnostic process and suggests the potential effectiveness of these models at the examination stage in antenatal clinics.
Our results are comparable to those of similar studies conducted by other authors [18, 19]. In these studies, a broader range of maternal factors was used as predictors, including specific parameters such as placental growth factor, soluble fms-like tyrosine kinase-1, pregnancy-associated plasma protein-A, and pulsatility index of the uterine arteries. One important aspect to consider when comparing these models is the gestational age range of the study cohort. For example, in the study by Marić I. et al. [18], pregnancies at similar stages were included, whereas in the study by Tan et al. [19], pregnancy periods were limited to 13+6 weeks. These collective findings suggest that the models we developed have substantial potential as an additional tool for the early screening of pregnant women. Given the demonstrated stability in external validation, these models could be considered for practical use in future prospective studies. To further enhance the accuracy metrics, an increase in the size of the training sample is recommended, particularly with regard to data related to the predicted class [20].
ML is an effective tool for developing models to predict and diagnose rare and multifactorial pathological processes such as PE. Routine clinical and laboratory parameters serve as factors that can be easily monitored and controlled during pregnancy. The creation of effective medical predictive models is a complex and multistage process that involves the collection, processing, and analysis of a substantial amount of information, as well as active collaboration between medical professionals, statisticians, and data specialists.
Conclusion
The metrics of the final models are consistent with those of the previously published models. The results of the external validation indicate the relative stability of the models when applied to new data. This, combined with the quality indicators, suggests their potential utility in real-world clinical practice. This study is our first experience in predicting pregnancy complications based on real-world clinical data. The quality of a predictive model is directly dependent on the data used and the statistical algorithms, which will need to be refined in future studies.
References
- English F.A., Kenny L.C., McCarthy F.P. Risk factors and effective management of preeclampsia. Integr. Blood Pressure Control. 2015; 8: 7-12. https://dx.doi.org/10.2147/IBPC.S50641.
- Jim B., Karumanchi S.A. Preeclampsia: pathogenesis, prevention, and long-term complications. Semin. Nephrol. 2017; 37(4): 386-97. https://dx.doi.org/10.1016/j.semnephrol.2017.05.011.
- Bartsch E., Medcalf K.E., Park A.L., Ray J.G.; High Risk of Pre-eclampsia Identification Group. Clinical risk factors for pre-eclampsia determined in early pregnancy: systematic review and meta-analysis of large cohort studies. BMJ. 2016; 353: i1753. https://dx.doi.org/10.1136/bmj.i1753.
- De Kat A.C., Hirst J., Woodward M., Kennedy S., Peters S.A. Prediction models for preeclampsia: a systematic review. Pregnancy Hypertens. 2019; 16: 48-66. https://dx.doi.org/10.1016/j.preghy.2019.03.005.
- Thangaratinam S., Allotey J., Marlin N., Dodds J., Cheong-See F., von Dadelszen P. et al. Prediction of complications in early-onset pre-eclampsia (PREP): development and external multinational validation of prognostic models. BMC Med. 2017; 15(1): 68. https://dx.doi.org/10.1186/s12916-017-0827-3.
- Zhang Y., Chen X.L., Chen W.M., Zhou H.B. Prognostic nomogram for the overall survival of patients with newly diagnosed multiple myeloma. Biomed. Res. Int. 2019; 2019: 5652935. https://dx.doi.org/10.1155/2019/5652935.
- Chen W., Sun S. Clinical application of a multiparameter-based nomogram model in predicting preeclampsia. Evid. Based Complement. Alternat. Med. 2022; 2022: 7484112. https://dx.doi.org/10.1155/2022/7484112.
- Moons K.G., Altman D.G., Reitsma J.B., Ioannidis J.P., Macaskill P., Steyerberg E. W. et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann. Intern. Med. 2015; 162(1): W1-73. https://dx.doi.org/10.7326/M14-0698.
- Ding Y., Simonoff J.S. An investigation of missing data methods for classification trees. Econometrics: Data Collection & Data Estimation Methodology eJournal. Publ. 1 December 2006. https://dx.doi.org/10.5555/1756006.1756012.
- Awais M., Shamshad F., Bae S. Towards an adversarially robust normalization approach. Cite as:arXiv:2006.11007 [cs.LG] (or arXiv:2006.11007v1 [cs.LG] for this version) . https://dx.doi.org/10.48550/arXiv.2006.11007.
- Лучинин А.С. Искусственный интеллект в гематологии. Клиническая онкогематология. Фундаментальные исследования и клиническая практика. 2022; 15(1): 16-27. [Luchinin A.S. Artificial Intelligence in Hematology. Clinical Oncohematology. 2022; 15(1): 16-27. (in Russian)]. https://dx.doi.org/10.21320/2500-2139-2022-15-1-16-27.
- Van Calster B., McLernon D.J., van Smeden M., Wynants L., Steyerberg E.W.; Topic Group ‘Evaluating diagnostic tests and prediction models’ of the STRATOS initiative. Calibration: the Achilles heel of predictive analytics. BMC Med. 2019; 17(1): 230. https://dx.doi.org/10.1186/s12916-019-1466-7.
- Zoubir A.M., Iskandler D.R. Bootstrap methods and applications. IEEE Signal Process. Mag. 2007; 24(4): 10-9. https://dx.doi.org/1010.1109/MSP.2007.4286560.
- Lundberg S.M., Erion G., Chen H., DeGrave A., Prutkin J.M., Nair B. et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020; 2(1): 56-67. https://dx.doi.org/10.1038/s42256-019-0138-9.
- Министерство здравоохранения Российской Федерации. Преэклампсия. Эклампсия. Отеки, протеинурия и гипертензивные расстройства во время беременности, в родах и послеродовом периоде. Клинические рекомендации. М.; 2021. 79с. [Ministry of Health of the Russian Federation. Preeclampsia. Eclampsia. Edema, proteinuria and hypertensive disorders during pregnancy, childbirth and the postpartum period. Clinical guidelines. Moscow; 2021. 79p. (in Russian)].
- Palomaki G.E., Haddow J.E., Haddow H.R., Salahuddin S., Geahchan C., Cerdeira A.S. et al. Modeling risk for severe adverse outcomes using angiogenic factor measurements in women with suspected preterm preeclampsia. Prenat. Diagn. 2015; 35(4): 386-93. https://dx.doi.org/10.1002/pd.4554.
- March M.I., Geahchan C., Wenger J., Raghuraman N., Berg A., Haddow H. et al. Circulating angiogenic factors and the risk of adverse outcomes among haitian women with preeclampsia. PloS One. 2015; 10(5): e0126815. https://dx.doi.org/10.1371/journal.pone.0126815.
- Marić I., Tsur A., Aghaeepour N., Montanari A., Stevenson D.K., Shaw G.M., Winn V.D. Early prediction of preeclampsia via machine learning Am. J. Obstet. Gynecol. MFM. 2020; 2(2): 100100. https://dx.doi.org/10.1016/j.ajogmf.2020.100100.
- Tan M.Y., Syngelaki A., Poon L.C., Rolnik D.L., O’Gorman N., Delgado J.L. et al. Screening for pre-eclampsia by maternal factors and biomarkers at 11-13 weeks’ gestation. Ultrasound Obstet. Gynecol. 2018; 52(2): 186-95. https://dx.doi.org/10.1002/uog.19112.
- Riley R.D., Debray T.P.A., Collins G.S., Archer L., Ensor J., van Smeden M., Snell K.I.E. Minimum sample size for external validation of a clinical prediction model with a binary outcome. Stat. Med. 2021; 40(19): 4230-51. https://dx.doi.org/10.1002/sim.9025.
Received 18.04.2023
Accepted 29.09.2023
About the Authors
Anna E. Andreychenko, PhD, Head of the Artificial Intelligence Department, K-SkAI, +7(916)321-25-70, aandreychenko@webiomed.ru, https://orcid.org/0000-0001-6359-0763, 17, Varkaus Embankment, Petrozavodsk, 185901, Russia.Alexander S. Luchinin, PhD, Senior Researcher, Department of Organization and Support of Scientific Research, Kirov Research Institute of Hematology and Blood Transfusion, Federal Medical Biological Agency of Russia, +7(919)506-87-86, luchinin@niigpk.ru, https://orcid.org/0000-0002-5016-210X,
84, Derendyaeva str., Kirov, 610027, Russia.
Alexander A. Ivshin, PhD, Associate Professor, Head of the Department of Obstetrics and Gynecology, Dermatovenerology of the Medical Institute, Petrozavodsk State University, +7(909)567-12-51, scipeople@mail.ru, https://orcid.org/0000-0001-7834-096X, 31, Krasnoarmeyskaya str., Petrozavodsk, 185035, Russia.
Andrey D. Ermak, PhD, Data analyst, Artificial Intelligence Department, K-SkAI, +7(977)563-52-72, aermak@webiomed.ru, https://orcid.org/0000-0002-0513-8557,
17, Varkaus Embankment, Petrozavodsk, 185901, Russia.
Roman E. Novitskiy, CEO, K-SkAI, +7(911)400-50-00, roman@webiomed.ru, https://orcid.org/0000-0002-2350-977X, 17, Varkaus Embankment, Petrozavodsk, 185901, Russia.
Alexander V. Gusev, PhD, Senior Researcher, Department of Scientific Fundamentals of Health Organization, Russian Research Institute or Health, +7(911)402-35-00,
agusev@webiomed.ai, https://orcid.org/0000-0002-7380-8460, 11, Dobrolyubova str., Moscow, 127254, Russia.
Corresponding author: Alexander A. Ivshin, scipeople@mail.ru



