Endometrial microbiome in women with and without a history of repeated failures of assisted reproductive technology: what are norm and pathology?
Aim. To investigate the endometrial microbiome in healthy fertile women and patients with multiple unsuccessful assisted reproduction technology (ART) cycles.Barinova V.V., Kuznetsova N.B., Bushtyreva I.O., Oksenyuk O.S., Dudurich V.V., Shatalov A.E.
Materials and methods. The study included 20 women with infertility and multiple unsuccessful ART cycles (group 1) and 15 healthy fertile women with a history of at least one full-term childbirth (group 2). The endometrial microbiome was analyzed using next-generation sequencing of the 16S rRNA gene.
Results. The endometrial microbiota of healthy fertile patients was dominated by Lactobacillus (29.4%), Comamonas (16.8%), and Mesorhizobium (6.0%). In infertile patients with multiple IVF failures, the most abundant were Lactobacillus (33.3%), Ralstonia (7.9%), and Pediococcus (4.8%). At the same time, the proportion of lactobacilli in patients of group 1 did not differ significantly from patients in group 2 (33.3% and 29.4%, respectively). Women with infertility and repeated unsuccessful IVF attempts had a significantly higher relative abundance of bacteria of the genera Brevundimonas and Ralstonia. At the same time, the fertile women in group 2 had a statistically significantly higher representation of Acidovorax, Brevibacillus, Caulobacter, Comamonas, Delftia, Distigma Pseudomonas, Schlegelella, Thermus.
Conclusion. The study findings confirm the notion that the uterus may indeed be a non-sterile compartment and the existence of the uterine microbiome as a whole. Lactobacilli are dominant members of the uterine microbiome but not absolutely dominant, i.e.,> 90%. At the same time, the proportion of lactobacilli in fertile patients and patients with multiple ART failures did not differ significantly.
Keywords
The endometrium has been traditionally considered sterile in non-pregnant, pregnant, and puerperal women, which has been proven by bacterial culture techniques [1, 2]. Indeed, endometrial infection or high concentrations of bacteria and its products such as endotoxin have been associated with implantation failures, spontaneous miscarriages, recurrent miscarriages, or spontaneous preterm birth [3–8].
At the same time, it is difficult to envision that a mucosa that is continuously exposed to microorganisms present in the lower genital tract and that is regularly invaded by sperm that can carry microorganisms into the endometrial cavity may be free of bacteria and its products.
The ascending infection from the vagina through the cervix to the uterus is considered the most likely route of endometrial colonization [9]. For example, radioactively labeled albumin spheres placed in the vagina ascend into the uterus as early as 2 minutes after instillation, suggesting that fluid and particles move between the vagina and uterus relatively freely [10]. Transfer of spermatozoa by a uterine peristaltic activity from the vagina to the uterus has been shown as a possible mechanism of endometrial microbial colonization.
On the other hand, the cervical mucus plug is partially impermeable to bacterial ascension from the vagina. Perhaps that is why there are data on a rapidly decreasing relative and absolute abundance of lactobacilli in the lower, middle, and upper third of the endometrium, respectively. The higher we move along the reproductive tract towards the fundus uteri and fallopian tubes, the less the presence of lactobacilli [9]. In summary, these data indicate that the endometrial cavity is not sterile in most women.
Recent studies in this area using molecular technologies have proven the presence of resident microflora in the human endometrium and its potential impact on women's reproductive health, especially on implantation and pregnancy outcomes [11–25].
The most widely debated hypothesis to date is the hypothesis of a Lactobacillus-dominated and non-Lactobacillus-dominated endometrial microbiota. Thus, it has been shown that Lactobacillus-dominated endometrial microbiota (>90% Lactobacillus spp.) is associated with successful implantation and a high live birth rate in patients undergoing IVF [15, 21]. In this regard, Logic dictates that an increase in the relative abundance of lactobacilli to 90% or more in women with non-Lactobacillus-dominated endometrial microbiota may contribute to an increase in implantation rates in infertile women and is associated with the concept of a healthy uterine microbiome.
Unfortunately, the current literature lacks sufficient evidence to form the concept of a normal and dysbiotic uterine microbiome, especially regarding the methods of its correction.
The present study aimed to investigate the endometrial microbiome in healthy fertile women with a history of at least one full-term childbirth and patients with multiple unsuccessful assisted reproduction technology (ART) cycles.
Materials and methods
The study included 35 women aged from 20 to 42 years who were managed at the Department of Obstetrics and Gynecology No. 1, Rostov State Medical University (Rostov-on-Don, Russia). The patients were divided into two groups.
Group 1 included 20 women with various forms of infertility and multiple (≥2) unsuccessful ART cycles. The exclusion criteria were any pathology of the uterus, including congenital malformations, fibroids, polyps, intrauterine synechiae, intrauterine manipulations within the last six months, the presence of an intrauterine device within the previous six months, any systemic inflammatory disease or severe non-gynecologic pathology, and the use of systemic antibacterial therapy in the last three months.
Group 2 consisted of 15 healthy fertile women without any complaints and complicated obstetric and gynecological history, and with a history of at least one full-term vaginal childbirth. In addition to the criteria for group 1, the exclusion criteria also included surgical abortions, self-abortions, missed miscarriage, postpartum dilation and curettage, and a history of cesarean section.
All patients signed informed consent to participate in the study. The study was reviewed and approved by the Research Ethics Committee of the Rostov State Medical University.
Sampling
All samples for the study were taken from the 22nd to the 24th day of the regular menstrual cycle. To minimize the risk of contaminating the endometrial samples in the vagina, a catheter with a hard outer part and an internal soft part used in embryo transfer were inserted into the uterine cavity, avoiding contact with the vaginal walls. After visualizing the cervix using a vaginal speculum, cervical mucus was removed by a sterile swab soaked in a chlorhexidine solution. Then, the catheter for embryo transfer was carefully inserted into the cervical canal without touching the vaginal walls. At the same time, the soft inner part of the catheter was inserted into the outer one, thus avoiding bacterial contamination of the cervical canal. After entering the uterine cavity, the inner catheter was gently pulled out from the outer end to the uterine fundus, and a sample was taken. After that, the inner catheter was again pulled back in the outer one, and the entire system was removed from the cervical canal and vagina. Finally, the endometrial sample was placed in an Eppendorf tube with a mucolytic transport medium (Central Research Institute of Epidemiology of Rospotrebnadzor, Moscow, Russia); the medium was kept at +4°C until DNA extraction.
Total DNA was isolated using the RIBO-prep nucleic acid extraction kit (Central Research Institute of Epidemiology of Rospotrebnadzor, Moscow, Russia) according to the manufacturer's instructions.
Libraries of 16S rRNA gene fragments were prepared according to Illumina's 16S Metagenomic Sequencing Library Preparation Workflow (Part No. 15044223 Rev. B). Five ng of total DNA was amplified for 25 cycles using the primers recommended in the protocol for the V3–V4 hypervariable region of the bacterial 16S rRNA gene. PCR amplification of the target region was performed using the KAPA HiFi Hot Start Ready Mix (2X) (Roche Diagnostics, Switzerland).
The resulting DNA fragments were purified using the AMPure XP paramagnetic beads (Beckman Coulter, USA). To introduce the multiplexing indexes and Illumina sequencing adapters, additional PCR amplification was conducted using the Nextera XT Index Kit (Illumina, USA) and the KAPA HiFi HotStart ReadyMix (2X) PCR mixture (Roche Diagnostics, Switzerland). The PCR product was then purified once again using the AMPure XP beads. After the library construction, the metagenomic sequencing was performed using the paired-end 2*151 bp Illumina MiSeq protocol (Illumina MiSeq, USA)
Data analysis
The obtained sequencing data were analyzed using the custom bioinformatic pipeline implemented in the R v.3.6 programming languages (R Core Team, 2014) and Python. At the first stage of the pipeline, the primer sequences were removed from the beginning of the readings. Reads that did not contain primers were also removed. Then, the last 25 nucleotides of reads were removed as having low quality, and the obtained data were processed using the DADA2 protocol to detect exact sequence variants [26]. After determining the exact variants of the sequences, forward and reverse readings were combined by concatenation, and the obtained sequences were used for Bayesian taxonomic classification [27] using the SILVA v132 reference database [28]. Species were identified using the DADA2 complete match algorithm using the SILVA v132 database sequences preprocessed with custom scripts to match the analysis.
Statistical analysis
The distribution of continuous variables was tested for normality using the Shapiro–Wilk test (Table 1). It was found that concentrations of the studied microorganisms were not normally distributed; therefore, they were reported as medians (Table 2). Comparing numerical data with non-normal distribution was performed with nonparametric Mann–Whitney tests. The differences were considered statistically significant at p <0.05. Calculations were performed in R (version 3.2, R Foundation for Statistical Computing, Vienna, Austria).
Results
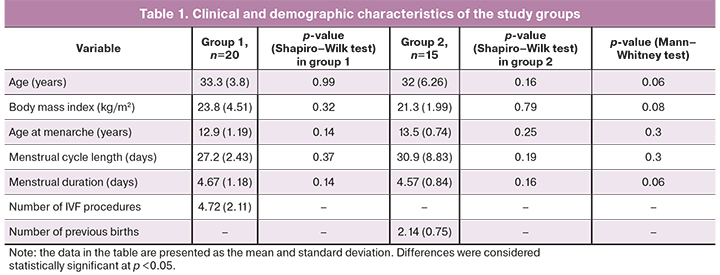
 The clinical and demographic characteristics of the patients of both groups are presented in Table 1. Patients of both groups were comparable in terms of body mass index, age at menarche, duration of the menstrual cycle length and menstrual duration, and age. These variables were normally distributed in both groups.
The clinical and demographic characteristics of the patients of both groups are presented in Table 1. Patients of both groups were comparable in terms of body mass index, age at menarche, duration of the menstrual cycle length and menstrual duration, and age. These variables were normally distributed in both groups.
Analysis of the endometrial microbiome using next-generation sequencing identified microorganisms belonging to a total of 19 phyla, 26 classes, 89 families, 257 genera, and 366 different species according to the taxonomic classification. In addition, comparisons between groups were made at the genus taxonomic level.
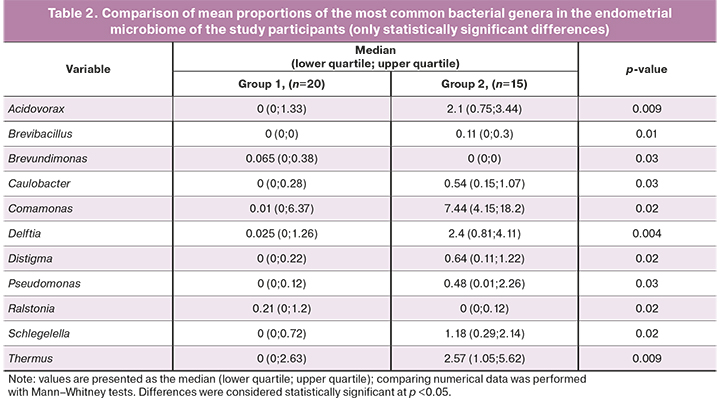
According to the genera level between the two groups, statistically, significant differences were revealed in the relative abundance of the following genera of microorganisms: Acidovorax, Brevibacillus, Brevundimonas, Caulobacter, Comamonas, Delftia, Distigma, Pseudomonas, Ralstonia, Schlegelella, and Thermus. Infertile women with repeated unsuccessful IVF attempts had statistically significantly higher proportions of bacterial genera Brevundimonas [0.065 (0; 0.38) in group 1 and 0 (0; 0) in group 2)] and Ralstonia [0.21 (0; 1.2) in group 1 and 0 (0; 0.12) in group 2].
At the same time, the fertile women had statistically significantly higher proportions of Acidovorax [0 (0; 1.33) in group 1 and 2.1 (0.75; 3.44) in group 2], Brevibacillus [0 (0; 0) in group 1 and 0.11 (0; 0.3) in group 2], Caulobacter [0 (0; 0.28) in group 1 and 0.54 (0.15; 1.07) in group 2, respectively], Comamonas [0.01 (0; 6.37) in group 1 and 7.44 (4.15; 18.2) group 2], Delftia [0.025 (0; 1.26) in group 1 and 2.4 (0.81; 4.11) in group 2], Distigma [0 (0; 0.22) in 1- group 1 and 0.64 (0.11; 1.22) in group 2, respectively], Pseudomonas [0 (0; 0.12) in group 1 and 0.48 (0.01; 2, 26) in group 2], Schlegelella [0 (0; 0.72) in group 1 and 1.18 (0.29; 2.14) in group 2], Thermus [0 (0; 2, 63) in group 1 and 2.57 (1.05; 5.62) in group 2] (Table 2).
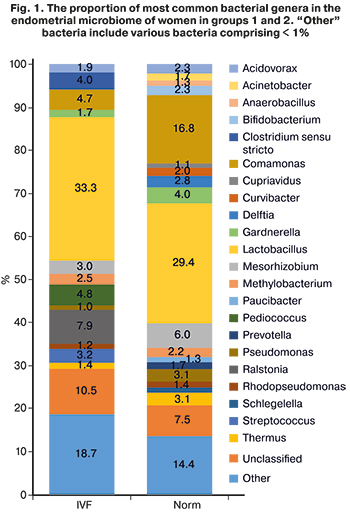
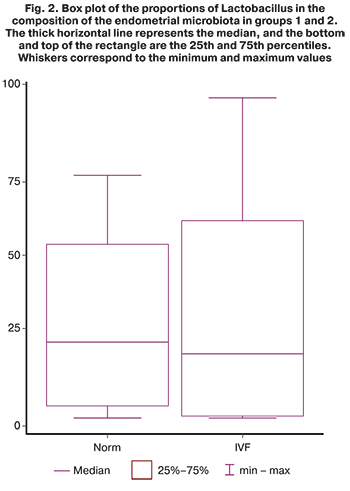 Analysis of the relative abundance of bacterial genera in the groups showed that among healthy fertile patients, the most prevalent were Lactobacillus (29.4%), Comamonas (16.8%) and Mesorhizobium (6.0%), and among infertile patients with multiple IVF failures Lactobacillus (33.3%), Ralstonia (7.9%) and Pediococcus (4.8%) (Fig. 1). At the same time, the proportion of lactobacilli in patients of group 1 did not differ significantly from the proportion of lactobacilli in patients of group 2 (33.3% and 29.4%, respectively) (Fig. 2). In addition, it is impossible to assert with confidence about the dominance of lactobacilli over 90% in the composition of the uterine microbiome in healthy women, as Moreno claims from his observations [15, 21]. The category "other microorganisms" included various microorganisms whose relative abundance in the microbiome was less than 1%. In patients of groups 1 and 2, "other" microorganisms accounted for 18.7% and 14.4%, respectively. Besides, unclassified microorganisms, those that could not be identified or whose genome is absent in available databases, accounted for 10.5% in group 1 and 7.5% in group 2. In general, it can be argued that the endometrial microbiome is sufficiently diverse, both in healthy fertile patients and in patients with multiple IVF failures, which may be directly related to the place of sampling for the study of the microbiome, namely, the uterine fundus. After all, the higher is the point of sampling in the female reproductive system, the lower the lactobacilli abundance. However, despite the diversity of microorganisms, there was almost complete absence (less than 1%) of the following bacterial genera: Streptococcus (3.2% in group 1), Ralstonia (7, 9% in group 1), Pediococcus (4.8% in group 1), and Clostridium (4.0% in group 1). At the same time, bacteria that were registered in fertile patients, but were not detected in significant proportions (more than 1%) in patients with infertility, were represented by the genera Acinetobacter (1.7% in group 2), Anaerobacillus (1.3% in group 2), Cupriavidus (1.1% in group 2), Curvibacter (2.0% in group 2), Delftia (2.8% in group 2), Paucibacter (1.3% in group 2), Prevotella (1.7% in group 2), and Shlegella (1.3% in group 2).
Analysis of the relative abundance of bacterial genera in the groups showed that among healthy fertile patients, the most prevalent were Lactobacillus (29.4%), Comamonas (16.8%) and Mesorhizobium (6.0%), and among infertile patients with multiple IVF failures Lactobacillus (33.3%), Ralstonia (7.9%) and Pediococcus (4.8%) (Fig. 1). At the same time, the proportion of lactobacilli in patients of group 1 did not differ significantly from the proportion of lactobacilli in patients of group 2 (33.3% and 29.4%, respectively) (Fig. 2). In addition, it is impossible to assert with confidence about the dominance of lactobacilli over 90% in the composition of the uterine microbiome in healthy women, as Moreno claims from his observations [15, 21]. The category "other microorganisms" included various microorganisms whose relative abundance in the microbiome was less than 1%. In patients of groups 1 and 2, "other" microorganisms accounted for 18.7% and 14.4%, respectively. Besides, unclassified microorganisms, those that could not be identified or whose genome is absent in available databases, accounted for 10.5% in group 1 and 7.5% in group 2. In general, it can be argued that the endometrial microbiome is sufficiently diverse, both in healthy fertile patients and in patients with multiple IVF failures, which may be directly related to the place of sampling for the study of the microbiome, namely, the uterine fundus. After all, the higher is the point of sampling in the female reproductive system, the lower the lactobacilli abundance. However, despite the diversity of microorganisms, there was almost complete absence (less than 1%) of the following bacterial genera: Streptococcus (3.2% in group 1), Ralstonia (7, 9% in group 1), Pediococcus (4.8% in group 1), and Clostridium (4.0% in group 1). At the same time, bacteria that were registered in fertile patients, but were not detected in significant proportions (more than 1%) in patients with infertility, were represented by the genera Acinetobacter (1.7% in group 2), Anaerobacillus (1.3% in group 2), Cupriavidus (1.1% in group 2), Curvibacter (2.0% in group 2), Delftia (2.8% in group 2), Paucibacter (1.3% in group 2), Prevotella (1.7% in group 2), and Shlegella (1.3% in group 2).
Discussion
Our study has some limitations. The first is the sample size of 20 patients in group 1 and 15 in group 2 that may not represent the population as a whole, but they may indicate some trend requiring further adequately powered study.
Another limitation of this study is the absence of negative controls. Some recent studies have reported that when applying sequence-based techniques to the study of microbiota present in low biomass environments, concurrent sequencing of negative control samples is strongly advised. If the endometrial microbiota does exist, then it is present in very low concentrations. Therefore, its molecular characteristics are influenced by background DNA contamination from extraction kits and reagents for PCR and sequencing [29]. For this reason, contaminating DNA can account for a significant part, if not all, of the recorded molecular microbial signatures within the endometrium. Therefore, it is essential that endometrial microbiota studies include technical controls for potential sources of background DNA contamination and a detailed presentation of the microbial profiles of these controls. Most of the studies available to date either do not include control samples or data on their microbial profile are insufficiently presented. When sequencing control samples of nutrient medium or air from the operating room, the number of reads should be hundreds of times less than the number of reads of material from the endometrium. Potential microbial contamination is possible from extraction kits, culture medium, air, which can significantly affect the reliability of the results, the interpretation of the clinical significance of the identified microorganisms. Therefore, the role of negative controls is of immense importance. However, this increases the cost of research by almost three times, which can complicate study planning.
The third limitation is a transcervical sampling that we used as the sample collection technique. Studies using transcervical sampling report higher rates of intrauterine bacterial colonization due to cervical or vaginal contamination of the endometrial specimens. They observed the presence of Lactobacillus microflora characteristic of the vaginal microbiome. At the same time, in studies using transabdominal sampling, the presence of microorganisms was more significant. Thus, Chen et al. used 16S rRNA sequencing to assess the microbiome of both the lower and upper sections of the female reproductive tract. For the upper sections, the average proportion of lactobacilli is only 1.7% (fallopian tubes). In the endometrium, the relative abundance of lactobacilli was 30.6%, but Acinetobacter (9.1%), Pseudomonas (9.1%), Vagococcus (7.3%), Sphingobium (5.0%), and Comamonadaceae (4.9%) were widely represented. [15]. Walther-Antonio et al. examined ten women after hysterectomy for benign neoplasms. In the myometrium and endometrium, Lactobacillus was found in extremely low proportions. The most represented in the endometrium were Shigella and Barnesiella [17]. In a study in which material was obtained from the middle third of the endometrium after hysterectomy, there was also no dominance of lactobacilli [30], and endometrial bacterial profiles were mainly represented by Acinetobacter, Pseudomonas, Comamonadaceae, and Cloacibacterium. Thus, in all studies using transcervical sampling, the predominance of lactobacilli in the composition of the uterine microbiome was reported, while with other sampling methods, the microbiome was more variable.
Our study has several distinct strengths. First, the findings confirm the concept of endometrial non-sterility and the existence of the uterine microbiome as a whole. Sequencing makes it possible to identify bacteria in the endometrium, but the analysis results do not allow for quantifying this presence; that is, it is impossible to say how many bacteria are found in a particular sample.
We obtained statistically significant differences between the groups for the following bacterial genera: Acidovorax, Brevibacillus, Brevundimonas, Caulobacter, Comamonas, Delftia, Distigma, Pseudomonas, Ralstonia, Schlegelella, and Thermus. However, their relative bacterial abundance in the composition of the uterine microbiome was often less than 1%; therefore, their influence on embryo implantation is questionable.
Our data confirm the current concept of lactobacilli dominance in the uterine microbiome. Considering the anatomical proximity of the uterine cavity and endometrium with the primary source of lactobacilli in the reproductive system - with the vagina - and the existence of uterine peristaltic activity, contributing to the capture of lactobacilli from the vagina and the ascending route of their entry into the uterine cavity, the dominance of lactobacilli becomes an understandable and logical fact. Various data on the percentage of lactobacilli in the microbiome can be explained by the place of sampling of the material for study: the closer to the region of the cervical canal, the higher the percentage of lactobacilli, the closer to the area of the uterine fundus, the lower this percentage.
In addition, our data refutes the role of the Lactobacillus-dominated and non-Lactobacillus-dominated endometrial microbiota in implantation failures. According to our data, the proportion of lactobacilli in fertile patients and patients with multiple ART failures did not differ significantly and amounted to 33.3% and 29.4%, respectively (Fig. 1). It is possible that this is due to the place of sampling. In our study, it was the uterine fundus, that is, the locus of the uterus, that is most distant from the source of lactobacilli (vagina). Besides, the relative abundance of lactobacilli over 50% in the microbiome was recorded in only 5 out of 15 women from the group of fertile patients and 7 out of 20 women from the group of women with ART failures (Fig. 3). The absolute dominance of lactobacilli – more than 90% – was noted in only three women among all study participants (35 people), which does not correlate with the data of Moreno [15]. In all likelihood, not only the relative abundance of lactobacilli affects the implantation success. It is necessary to search for pathogens triggering a cascade of inflammatory reactions in the endometrium, thereby determining the implantation failure. It is essential to study the endometrial microbiome concurrently with markers of chronic endometritis and histological findings of the endometrial samples.
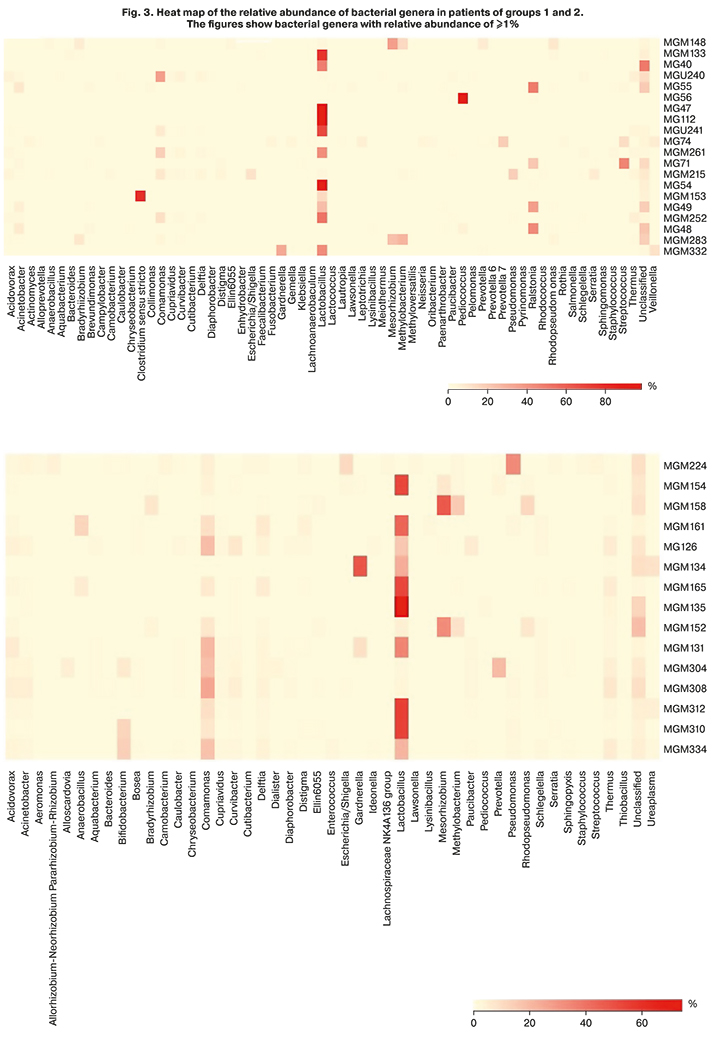
According to our data, women with infertility and repeated unsuccessful IVF attempts had statistically significantly higher relative proportions of bacterial genera Brevundimonas and Ralstonia. Fertile women in group 2 had statistically significantly higher proportions of Acidovorax, Brevibacillus, Caulobacter, Comamonas, Delftia, Distigma, Pseudomonas, Schlegelella, and Thermus. Whether this statistically significant difference is of clinical significance remains unclear since, first, the mean relative abundance of these bacterial genera was less than 1%. Secondly, all these microorganisms are opportunistic pathogens that may cause disease only under certain circumstances.
The most represented in healthy fertile patients were Lactobacillus, Comamonas, and Mesorhizobium, and in the group of infertile patients with multiple IVF failures – Lactobacillus, Ralstonia, and Pediococcus. Regarding Lactobacillus, their biological role in the composition of the microbiota is more or less clear. As for Comamonas, this genus belongs to opportunistic pathogens, resistant in the external environment, and characteristic of the gastrointestinal tract [31]. Mesorhizobium are also opportunistic microorganisms that occur in nature as a symbiont of plants, but there are isolated reports of their involvement in the development of peritonitis [32]. Ralstonia is generally characteristic of such an econiche as water. They can survive in conditions of low nutrient supply. In humans, they are involved in the development of osteomyelitis and meningitis [33]. Pediococcus is a genus of lactic acid-producing bacteria that are widely described as probiotics. The role of all these bacterial genera in the composition of the uterine microbiome remains insufficiently studied [34].
Conclusion
These study findings support the concept that the endometrium might not be as sterile as thought. Of course, the endometrium is not such a densely populated biotope as, for example, the intestinal lumen. Still, one cannot ignore the role of detected microorganisms in the etiology of various gynecological diseases and obstetric complications.
Our data confirm the theory of lactobacilli dominance in the composition of the endometrial microbiome. However, the proportion of lactobacilli did not differ significantly in healthy patients and patients with ART failures. Therefore, lactobacilli dominance cannot be considered as a factor determining the absolute success of implantation.
The mechanisms underlying successful implantation are more subtle and include, in addition to the influence of certain pathogenic microorganisms, also the local immune response of the endometrium. In this regard, it is essential to study the composition of the endometrial microbiome concurrently with markers of endometritis, histological findings of the endometrial samples, and spiral artery Doppler parameters and include them in diagnostic algorithms to evaluate the functional state of the endometrium.
Besides, knowledge about the composition of the endometrial microbiome entails the need to develop algorithms for correcting dysbiosis. In this case, it is imperative to consider the pathways of microorganisms entering the uterine cavity, including those ascending from the vagina, hematogenous from the intestines, and the oral cavity. All this opens up prospects for further studies.
References
- Butler B. Value of endometrial cultures in sterility investigation. Fertil. Steril. 1958; 9(3): 269-73. https://dx.doi.org/10.1016/s0015-0282(16)33070-9.
- Bollinger C.C. Bacterial flora of the nonpregnant uterus: a new culture technic. Obstet. Gynecol. 1964; 23: 251-5.
- Drbohlav P., Hálková E., Masata J., Rezácová J., Cerný V., Rossová D. The effect of endometrial infection on embryo implantation in the IVF and ET program. Ceska Gynekol. 1998; 63(3): 181-5. https://dx.doi.org/10.1093/humrep/deu292.
- Cicinelli E., Matteo M., Tinelli R., Lepera A., Alfonso R., Indraccolo U. et al. Prevalence of chronic endometritis in repeated unexplained implantation failure and the IVF success rate after antibiotic therapy. Hum. Reprod. 2015; 30(2): 323-30. https://dx.doi.org/10.1093/humrep/deu292.
- Cicinelli E., Matteo M., Tinelli R., Pinto V., Marinaccio M., Indraccolo U. et al. Chronic endometritis due to common bacteria is prevalent in women with recurrent miscarriage as confirmed by improved pregnancy outcome after antibiotic treatment. Reprod. Sci. 2014; 21(5): 640-7. https://dx.doi.org/10.1177/1933719113508817.
- Гусейнова Г.Э., Ходжаева З.С., Муравьева В.В. Роль микробиоты влагалища при досрочном преждевременном разрыве плодных оболочек. Акушерство и гинекология. 2020; 1: 20-5. [Guseinova G.E., Khodzhaeva Z.S., Muravyeva V.V. Role of the vaginal microbiota in preterm premature rupture of membranes. Obstetrics and gynecology. 2020; 1: 20-5. (in Russian)]. https://dx.doi.org/10.18565/aig.2020.1.20-25.
- Ходжаева З.С., Горина К.А., Тимошина И.В., Припутневич Т.В. Программирование здоровья новорожденного – роль материнского микробиома. Акушерство и гинекология: новости, мнения, обучение. 2019; 7(4): 61-5. [Khodzhaeva Z.S., Gorina K.A., Timoshina I.V., Priputnevich T.V. Programming of newborn health – the role of the maternal microbiome. Obstetrics and gynecology: news, opinions, training. 2019; 7(4): 61-5. (in Russian)]. https://dx.doi.org/10.24411/2303-9698-2019-14004.
- Кузнецова Н.Б., Буштырева И.О., Дыбова В.С., Баринова В.В., Полев Д.Е., Асеев М.В., Дудурич В.В. Микробиом влагалища у беременных с преждевременным разрывом плодных оболочек в сроке от 22 до 28 недель беременности. Акушерство и гинекология. 2021; 1: 94-102. [Kuznetsova N.B., Bushtyreva I.O., Dybova V.S., Barinova V.V., Polev D.E., Aseev M.V., Dudurich V.V. Vaginal microbiome in pregnant women with preterm prelabor rupture of membranes at 22–28 weeks’ gestation. Obstetrics and gynecology. 2021; 1: 94-102. (in Russian)]. https://dx.doi.org/10.18565/aig.2021.1.94-102.
- Mitchell C.M., Haick A., Nkwopara E., Garcia R., Rendi M., Agnew K. et al. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am. J. Obstet. Gynecol. 2015; 212(5): 611.e1-9. https://dx.doi.org/10.1016/j.ajog.2014.11.043.
- Zervomanolakis I., Ott H.W., Hadziomerovic D., Mattle V., Seeber B.E., Virgolini I. et al. Physiology of upward transport in the human female genital tract. Ann. N. Y. Acad. Sci. 2007; 1101: 1-20. https://dx.doi.org/10.1196/annals.1389.032.
- Marchesi J.R., Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015; 3: 31. https://dx.doi.org/10.1186/s40168-015-0094-5.
- Fang R.L., Chen L.X., Shu W.S., Yao S.Z., Wang S.W., Chen Y.Q. Barcoded sequencing reveals diverse intrauterine microbiomes in patients suffering with endometrial polyps. Am. J. Transl. Res. 2016; 8(3): 1581-92.
- Franasiak J.M., Werner M.D., Juneau C.R., Tao X., Landis J., Zhan Y. et al. Endometrial microbiome at the time of embryo transfer: next-generation sequencing of the 16S ribosomal subunit. J. Assist. Reprod. Genet. 2016; 33(1): 129-36. https://dx.doi.org/10.1007/s10815-015-0614-z.
- Khan K.N., Fujishita A., Masumoto H., Muto H., Kitajima M., Masuzaki H., Kitawaki J. Molecular detection of intrauterine microbial colonization in women with endometriosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016; 199: 69-75. https://dx.doi.org/10.1016/j.ejogrb.2016.01.040.
- Moreno I., Codoñer F.M., Vilella F., Valbuena D., Martinez-Blanch J.F., Jimenez-Almazán J. et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am. J. Obstet. Gynecol. 2016; 215(6): 684-703. https://dx.doi.org/10.1016/j.ajog.2016.09.075.
- Verstraelen H., Vilchez-Vargas R., Desimpel F., Jauregui R., Vankeirsbilck N., Weyers S. et al. Characterization of the human uterine microbiome in non-pregnant women through deep sequencing of the V1-2 region of the 16S rRNA gene. Peer J. 2016; 4: e1602. https://dx.doi.org/10.7717/peerj.
- Walther-Antonio M., Chen J., Multinu F., Hokenstad A., Distad T.J., Cheek E.H. et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016; 8(1): 122. https://dx.doi.org/10.1186/s13073-016-0368-y.
- Chen C., Song X., Wei W., Zhong H., Dai J., Lan Z. et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 2017; 8(1): 875. https://dx.doi.org/10.1038/s41467-017-00901-0.
- Miles S.M., Hardy B.L., Merrell D.S. Investigation of the microbiota of the reproductive tract in women undergoing a total hysterectomy and bilateral salpingo-oopherectomy. Fertil. Steril. 2017; 107(3): 813-20. https://dx.doi.org/10.1016/j.fertnstert.2016.11.028.
- Tao X., Franasiak J.M., Zhan Y., Scott R.T. III, Rajchel J., Bedard J. et al. Characterizing the endometrial microbiome by analyzing the ultra-low bacteria from embryo transfer catheter tips in IVF cycles: Next generation sequencing (NGS) analysis of the 16S ribosomal gene. Hum. Microbiome J. 2017; 3(1): 15-21. https://dx.doi.org/10.1016/j.humic.2017.01.004.
- Kyono K., Hashimoto T., Nagai Y., Sakuraba Y. Analysis of endometrial microbiota by 16S ribosomal RNA gene sequencing among infertile patients: a single-center pilot study. Reprod. Med. Biol. 2018; 17(3): 297-306. https://dx.doi.org/10.1002/rmb2.12105.
- Liu Y., Wong K.K., Ko E.Y., Chen X., Huang J., Tsui S.K. et al. Systematic comparison of bacterial colonization of endometrial tissue and fluid samples in recurrent miscarriage patients: implications for future endometrial microbiome studies. Clin. Chem. 2018; 64(12): 1743-52. https://dx.doi.org/10.1373/clinchem.2018.289306.
- Pelzer E.S., Willner D., Buttini M., Huygens F. A role for the endometrial microbiome in dysfunctional menstrual bleeding. Antonie Van Leeuwenhoek. 2018; 111(6): 933-43. https://dx.doi.org/10.1007/s10482-017-0992-6.
- Wee B.A., Thomas M., Sweeney E.L., Frentiu F.D., Samios M., Ravel J. et al. A retrospective pilot study to determine whether the reproductive tract microbiota differs between women with a history of infertility and fertile women. Aust. N. Z. J. Obstet. Gynaecol. 2018; 58(3): 341-8. https://dx.doi.org/10.1111/ajo.12754.
- Кебурия Л.К., Смольникова В.Ю., Припутневич Т.В., Муравьева В.В. Микробиота полости матки и ее влияние на репродуктивные исходы. Акушерство и г инекология. 2019; 2: 22-7. [Keburia L.K., Smolnikova V.Yu., Priputnevich T.V., Muravyeva V.V. Uterine microbiota and its effect on reproductive outcomes. Obstetrics and gynecology. 2019; 2: 22-7. (in Russian)]. https://dx.doi.org/10.18565/aig.2019.2.22-27.
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016; 13(7): 581-3. https://dx.doi.org/10.1038/nmeth.3869.
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007; 73(16): 5261-7. https://dx.doi.org/10.1128/AEM.00062-07.
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P. et al. The SILVA ribosomal RNA.gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013; 41(Database issue): D590-6. https://dx.doi.org/10.1093/nar/gks1219.
- Salter S.J., Cox M.J., Turek E.M., Calus S.T., Cookson W.O., Moffatt M.F. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014; 12: 87. https://dx.doi.org/10.1186/s12915-014-0087-z.
- Winters A.D., Romero R., Gervasi M.T., Gomez-Lopez N., Tran M.R., Garcia-Flores V. Does the endometrial cavity have a molecular microbial signature? Sci. Rep. 2019; 9(1): 9905. https://doi.org/10.1038/s41598-019-46173-0.
- Khalki H., Deham H., Taghouti A., Yahyaoui G., Mahmoud M. Appendicite à comamonas testosteroni [Comamonas testosteroni appendicitis]. Med. Mal. Infect. 2016; 46(3): 168-70. https://dx.doi.org/10.1016/j.medmal.2015.12.009.
- Krishnan A., Raby E., Puttagunta H., Leung M., Thomas M. Mesorhizobium peritonitis: The first reported case. Nephrology. 2016; 22(1): 96. https://dx.doi.org/10.1111/nep.12716.
- Ryan M.P., Adley C.C. Ralstonia spp.: emerging global opportunistic pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2014; 33(3): 291-304. https://dx.doi.org/10.1007/s10096-013-1975-9.
- Porto M.C., Kuniyoshi T.M., Azevedo P.O., Vitolo M., Oliveira R.P. Pediococcus spp.: An important genus of lactic acid bacteria and pediocin producers. Biotechnol. Adv. 2017; 35(3): 361-74. https://dx.doi.org/10.1016/j.biotechadv.2017.03.004.
Received 02.12.2020
Accepted 30.04.2021
About the Authors
Viktoriya V. Barinova, PhD, Teaching Assistant at the Department of Obstetrics and Gynecology No. 1, Rostov State Medical University, Ministry of Health of theRussian Federation; Head of the Obstetric Department of the Clinic of Professor Bushtyreva LLC. Tel.: +7(928)909-55-68. E-mail: victoria-barinova@yandex.ru.
ORCID ID: 0000-0002-8584-7096, WoS Researcher ID: AAH-3314-2019, Scopus Author ID: 2578513.
344022, Russia, Rostov-on-Don, Nakhichevanskiy str., 29; 344011, Russia, Rostov-on-Don, Soborniy str., 58/7.
Natalya B. Kuznetsova, Dr. Med. Sci., Professor at the Center for Simulation Training, Rostov State Medical University, Ministry of Health of the Russian Federation;
Chief Physician of the Clinic of Professor Bushtyreva LLC. Tel.: +7(928)770-97-62. E-mail: lauranb@inbox.ru.
344022, Russia, Rostov-on-Don, Nakhichevanskiy str., 29; 344011, Russia, Rostov-on-Don, Sobornyi str., 58/7.
Irina O. Bushtyreva, Dr. Med. Sci., Professor, Director of the Clinic of Professor Bushtyreva LLC. Tel.: +7(928)296-15-97. E-mail: kio4@mail.ru.
344011, Russia, Rostov-on-Don, Sobornyi str., 58/7.
Oksana S. Oksenyuk, PhD, Head of the Central Research Laboratory, Rostov State Medical University, Ministry of Health of the Russian Federation. Tel.: +7(928)100-94-34. E-mail: oksenuk_o@bk.ru. ORCID ID: 0000-0003-2592-1167, Scopus Author ID: 57194872826, Researcher ID: Q-5263-2017.
344022, Russia, Rostov-on-Don, Nakhichevanskiy str., 29.
Vasilisa V. Dudurich, Biologist-Geneticist, Director for Development of the Medical Genetic Center “Serbalab”. Tel.: +7(914)542-20-10. E-mail: vdudurich@cerbalab.ru.
199106, Russia, St. Petersburg, V.O., Bolshoy ave., 90-2.
Alexander E. Shatalov, 6th year student at the Medical and Preventive Faculty, Rostov State Medical University, Ministry of Health of the Russian Federation.
Tel.: +7(928)191-35-82. E-mail: shatal321@mail.ru. 344022, Russia, Rostov-on-Don, Nakhichevanskiy str., 29.
For citation: Barinova V.V., Kuznetsova N.B., Bushtyreva I.O., Oksenyuk O.S., Dudurich V.V., Shatalov A.E. Endometrial microbiome in women with and without a history of repeated failures of assisted reproductive technology: what are norm and pathology?
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2021; 6: 105-114 (in Russian)
https://dx.doi.org/10.18565/aig.2021.6.105-114



