Expression of exosomal microRNA in high-grade ovarian cancer and ovarian endometriotic cysts
Recently, exosomal microRNA has been considered a potential noninvasive biomarker of various proliferative processes, particularly affecting the female reproductive system.Iurova M.V., Eldarov Ch.M., Bobrov M.Yu., Khabas G.N., Pavlovich S.V.
Objective: To investigate the potential of plasma exosomal microRNA expression in patients with serous ovarian cancer, extragenital endometriosis, and ovarian endometriotic cysts (OEC) as potential markers of these diseases. Materials and methods: The study included seven patients with histologically confirmed high-grade serous ovarian cancer, six women in a control group without this pathology ("Control-ovarian cancer"), six patients with OEC, and five women of reproductive age without this pathology ("Control-OEC"). The exosomes were isolated from the blood, and the composition of plasma-derived exosomal microRNAs was determined using next¬generation sequencing.
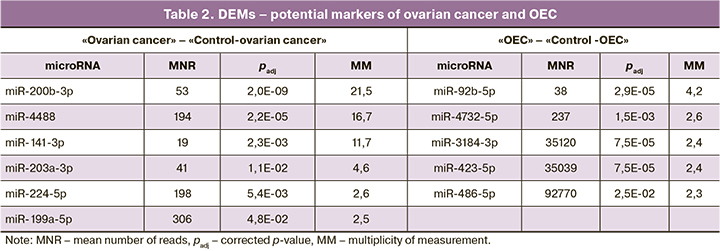
Results: Pairwise comparison of miRNA content in the groups "ovarian cancer" — "Control-ovarian cancer" and "OEC"— "Control-OEC" identified 22 and 13 differentially expressed miRNAs (DEM) with statistically significant 2-fold or more differences. Potential markers for ovarian cancer included miR-141-3p, -199a-5p, -200b-3p, -203a-3p, -224-5p, and -4488 andfor OEC, miR-92b-5p, -486-5p, -3184-3p, -4732-5p, and -423¬5p. Potential target genes were identifiedfor DEM in each pair of groups. The intracellular signaling pathways most likely involved in the pathogenesis of ovarian cancer and OEC were searched based on their lists. Potential target genes of several signaling pathways regulated by one or more DEMs in ovarian cancer and OEC included AKT1, ATM, BARD1, BAX, BCL2, BRCA1, CASP3, CDK4, CHEK1, CHEK2, JAK1, MDM2, PLK1, PTEN, RB1, SMAD2, SMAD3, and TP53. Functional clustering of target genes showed their involvement in regulating signalingpathways ofgrowth factors, cell cycle, apoptosis, and DNA repair.
Conclusion: The detection of unique DEMs in plasma exosomes indicates the presence of specific changes in the microRNA profile characteristic of ovarian cancer and OEC. The listed exosomal microRNAs can be considered candidates for markers of the studied proliferative processes. Multiple interactions between the identified microRNA target genes indicate their significant contribution and joint involvement into proliferative processes in ovarian cancer and OEC.
Keywords
The diagnosis of pathological proliferative processes of female reproductive organs is a challenging issue in modern gynecology. Ovarian cancer is the fifth leading cause of all cancer-related deaths among women [1]. Endometriosis is characterized by the presence of endometrial-like tissue outside the uterus and affects about 10–15% of women of reproductive age [2, 3], causing infertility and significantly worsening patients' quality of life [4]. Endometriosis is considered a benign estrogen-dependent inflammatory disorder. However, endometriotic heterotopias show similar characteristics to cells undergoing malignant transformation, including impaired cell proliferation, apoptosis, migration, the ability to stimulate angiogenesis, resistance to immunocompetent cells leading to growth and spread of endometriotic foci [5].
Circulating microRNAs are a subclass of small non-coding RNAs that have been found in almost all body fluids, where they are believed to be involved in intercellular communications [6]. The biogenesis of microRNAs in the cell occurs first in the nucleus and then in the cytoplasm, where they provide post-transcriptional regulation of target gene expression [7]. Changes in microRNA expression have been demonstrated in ovarian tumor tissues and in ovarian endometriosis in several studies, indicating their essential role in the pathogenesis of these diseases [8, 9]. MicroRNAs can be secreted in a complex with specific proteins or as part of exosomes into the intercellular space or circulatory channel [10]. Exosomes are extracellular vesicles generated by all cells containing a specific molecular repertoire that regulates the cellular behavior of target cells [10, 11]. Changes in circulating exosome composition, especially microRNA content, can reflect the activity of a pathological process, and in this regard exosomal microRNAs are considered promising non-invasive markers [12]. Many studies have investigated the diagnostic potential of circulating microRNAs in different types of ovarian cancer [13]. The results turned out to be contradictory, which can be explained by various reasons, including differences in the study design, the use of serum or plasma, and the methods of microRNA detection and identification. Besides, the vast majority of studies analyzed the total composition of plasma or serum microRNA without individual analysis of exosomal fractions. It should be noted that the composition of circulating exosomal microRNAs in endometriosis remains unstudied. Another limitation of studies in this field is the use of polymerase chain reaction, which, although being highly sensitive in detecting specific microRNAs, does not allow evaluating the entire spectrum of microRNAs in the studied samples. Therefore, in this work we used next-generation sequencing, which allows for the most complete profiling of exosomal microRNAs.
The present study aimed to investigate the differential expression of plasma exosomal microRNA in patients with advanced serous ovarian cancer and extragenital endometriosis as potential markers of these diseases and in control subjects.
Materials and methods
This cross-sectional study was conducted at the V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia. The study included seven patients with histologically confirmed high-grade stages III–IV serous ovarian cancer according to the classification of FIGO (The International Federation of Gynecology and Obstetrics) and six patients with stage III-IV OEC according to the revised American Society of Reproductive Medicine (rASRM) staging classification of endometriosis.
The diagnosis was verified histologically. Blood samples were taken from women of comparable postmenopausal age [(n=6, 65 (10) years in the control group and 63 (5) years in the group of patients with ovarian cancer, p=0.59)] and reproductive age [(n=5, 34 (2) years in the control group and 31 (1) year in patients with OEC, p=0.06)].
Inclusion criteria were informed consent to participate in the study, data on concomitant extragenital pathology (including acute inflammatory and autoimmune processes), benign and malignant diseases of any location at the time of inclusion in the study and suffering earlier, follicular (proliferative) phase of menstrual cycle for women of reproductive age, postmenopause for patients with ovarian cancer and women in the control group, no hormonal medications. Reproductive organ diseases and extragenital pathology were excluded from the control groups based on anamnesis, pelvic ultrasound examination, clinical blood test, and CA125 and HE4 oncomarkers.
Patients' blood samples (10 ml) were collected before surgical intervention. Blood was transported to the laboratory in tubes with EDTA (Ethylenediaminetetraacetic acid) within 30 minutes after collection. Blood was centrifuged at 300×g during 20 min at 4°C. After centrifugation, the supernatant was withdrawn and centrifuged again at 3000 ×g during 15 minutes. The resulting plasma was introduced into cryovials in 1.5–2 ml portions. The labeled samples were frozen and stored at -80°C. Plasma samples were prepared and stored in the Laboratory for Biomaterials Collection and Storage at the V.I. Kulakov NMRC for OG&P.
To identify potential markers of ovarian cancer and OEC, the representation in exosomes of different patient groups was examined by high-throughput sequencing. Before RNA isolation, blood plasma was thawed and centrifuged at 16000 g, at 4°C. Exosomal RNA was isolated from 3 ml of plasma obtained after centrifugation using the ExoRNeasy Maxi kit (Qiagen) and then used to create complementary small RNA DNA libraries using the NEBNext Multiplex Small RNA Library Prep Set for Illumina, in strict accordance with the manufacturer's procedures. MicroRNAs were analyzed by high-throughput sequencing on the Illumina NextSeq platform, using the Illumina NextSeq 500/500 High Output Kit ver. 2.5 (75 cycles).
Sequencing yielded nucleotide sequence reads for the samples, which were mapped to the miRbase v21 database (miRbase.org) to identify mature microRNAs. The DESeq2 algorithm [14] was used to normalize the data and analyze the differential expression of microRNAs in the groups. MicroRNAs with the number of reads ≥10 were selected for further study; the multiplicity of changes ≥1.5 was also considered. When selecting microRNAs, results were considered statistically significant if padj<0.05, where padj is the p-value with correction for the average false discovery rate (FDR) for multiple comparisons. For selected microRNAs, we searched for possible targets in the microRNA-target interactions database (miRTarBase [15]). The database contains information on over fifty thousand microRNA-target interactions, data on which are regularly updated according to a systematic analysis of relevant studies. Only microRNA-gene interactions that were detected by one or more relevant methods (‘strong’ according to the database authors) were selected: real-time polymerase chain reaction (qPCR), WesternBlot, and high-throughput reporter assay kit. The resulting lists of potential targets were used to search for signaling pathways regulated by these targets and, as a consequence, change their activity. A model of known and predicted protein-protein interactions was used (database https://string-db.org/) and the representation of target genes in various signaling pathways of WikiPatways (https://www.wikipathways.org) was analyzed. As a result of the enrichment of signaling pathways for each pair of compared groups, those were selected that showed the highest statistical significance (XD score, q-value), determined by Fisher's exact test of overlapping with the given list of targets. Corrections for tissue specificity were used in the selection of signaling pathways. The pathways with the highest representation indicating activity in pathological processes in the ovaries were selected.
Results
Sequencing data identified 480 microRNAs in exosomes, with varying representation in the study groups. Lists of differentially expressed microRNAs (DEMs), and the multiplicity and direction of changes, are shown in Table 1 and Figure 1.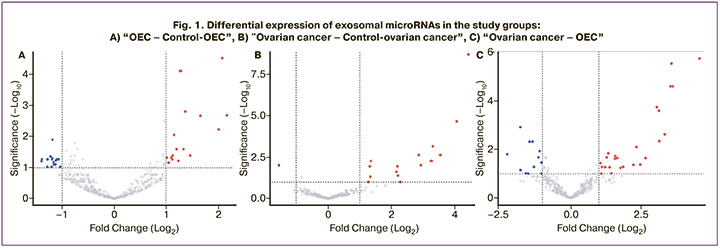
Among the identified DEMs that differentiated patients in the reproductive age control group (Fig. 1A), 22 showed a multiplication by factor of two or more in OEC: 14 DEMs with increased and 8 DEMs with decreased expression. For ovarian cancer, statistically significant differences were obtained in the expression of 13 microRNAs (12 DEMs increased and 1 DEM decreased), which differentiated patients from postmenopausal control subjects (Figure 1B); the highest multiplicity of changes was observed for miR-200b-3p, miR-4488, miR-141-3p (21.5, 16.7 and 11.7 times increase, respectively). Differences in microRNA representation in blood exosomes of patients with ovarian cancer and OEC were also evaluated. Twenty-seven DEMs were identified, of which 18 were overexpressed and 9 were underexpressed by a factor of two or more, respectively (Table 1, Fig. 1B).
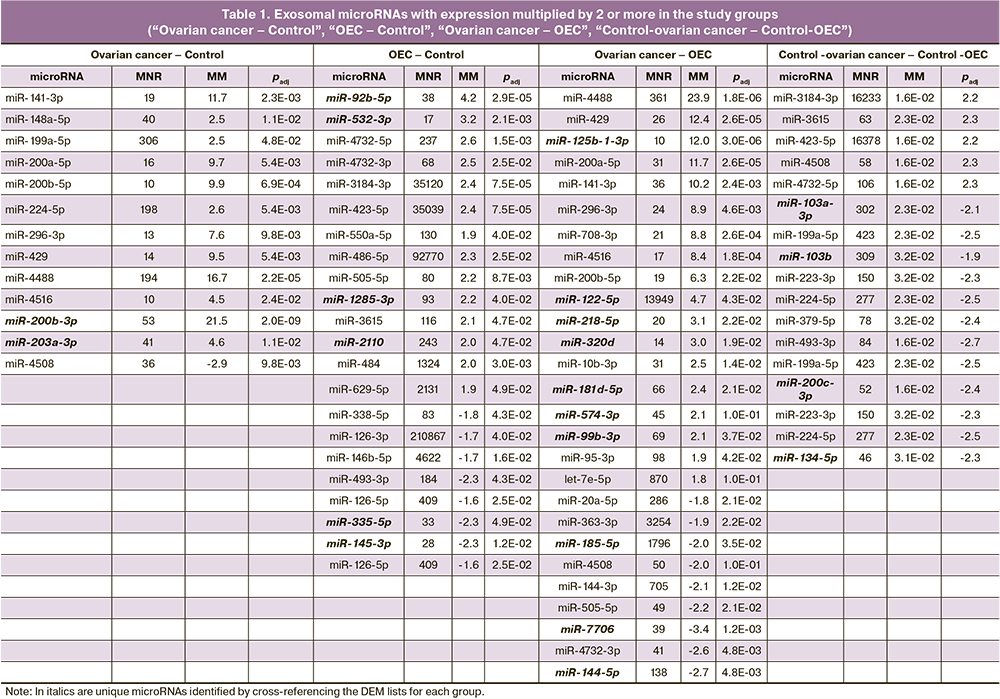
Since the study groups were represented by patients of different ages, a comparison between the control groups was also performed to assess the possible influence of age. Significant differences in the representation of 17 exosomal microRNAs were found in women of the reproductive and postmenopausal control groups: An increase of 5 DEM and a decrease of 12 DEM were found in postmenopausal women. However, the cross-referencing of the DEM lists of the ovarian cancer-OEC and control-ovarian cancer-OEC comparison groups showed the presence of only two common DEMs (miR-484 and miR-4508). Furthermore, their expression was reduced in ovarian cancer and OEC compared to the control group. This indicates that there was no influence of age on the representation of the other microRNAs in the ovarian cancer – OEC comparison group (Fig. 2).
The cross-referencing of the DEM lists obtained in the “OEC” – ”Control-OEC”, “ovarian cancer” – "Control-ovarian cancer” comparisons showed no common microRNAs (Fig. 2), indicating that the microRNA set for each pathology is unique. The microRNA lists for each intersection are presented in Table 1. This cross-referencing also resulted in unique DEMs with a multiple of two or more changes for each group: 18 microRNAs for OEC, 2 microRNAs for ovarian cancer, and 26 microRNAs for “ovarian cancer – OEC” (highlighted in italics in Table 1). The presence of unique DEMs in blood plasma exosomes may indicate specific changes in their composition depending on the pathology under investigation. The DEMs selected as potential markers are presented in Table 2.
In addition to unique DEMs, DEMs that occurred in the “ovarian cancer – OEC” and “Control-ovarian cancer” – “Control-OEC” comparison pairs were added to the presented lists. Thus, for ovarian cancer, the DEM markers miR-199a-5p and miR-224-5p were also detected when comparing control groups of different ages. Given the prevalence of ovarian cancer predominantly in postmenopausal age, using these DEMs as markers will not lead to false-positive results. However, the age factor must be considered in their potential use.
The DEM lists obtained during the study were used to search for potential target genes in the MirTarBase v. 7.0. Since microRNAs can theoretically realize interactions with numerous matrix RNAs (mRNAs), only the most likely microRNA-mRNA pairs whose interactions were found to be highly validated (confirmed simultaneously by qPCR, WesternBlot, reporter assay kit, etc.), using MirTarBase data) were selected from the search results. The target gene sets thus obtained were used for gene set enrichment analysis (gene set enrichment analysis) using the WikiPathways database. Based on the statistical parameters of significance and validity of the selection of target gene sets for a particular process (XD, q value [16]) and their representation for each comparison group, pathways were selected from which the lists of the most represented in the two comparison groups were selected. The XD parameter indicates how significantly the gene list under study is integrated into a particular process implemented by a particular network of molecular interactions. The parameter q represents the corrected validity value (p) for the XD parameter. The selected pathways for the comparison groups are presented in Table 3.
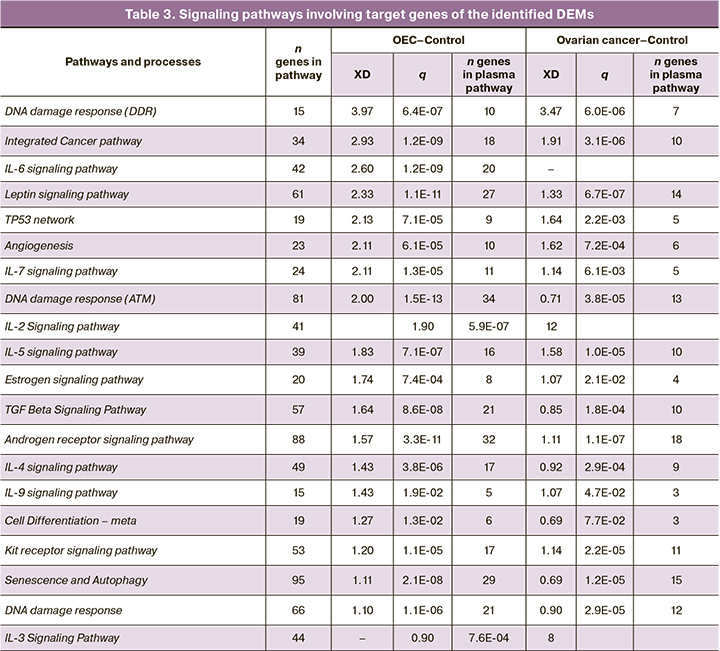
It is interesting to note cellular processes and signaling cascades, pathways activated by estrogens and androgens, growth factors, and morphogenic regulators. We have also identified a group of pathways controlled by interleukins (IL-2, -3, -4, -5, -6, -7, -9), which may participate in the regulation of inflammatory processes during the formation of pathologic foci in cancer and endometriosis. Intracellular processes involved in the regulation of DNA repair in the event of DNA damage were also established. Of note is the enrichment with high values of XD parameter by regulatory pathways associated with DNA damage, implemented with the participation of microRNA (miRNA involved in DDR). Activation of these pathways as well as signaling mediated by interleukins, TP53 and growth factors may indicate the development of an inflammatory process and oxidative stress at the sites of exosome formation studied. DNA damage, in turn, may be a factor inducing the processes of apoptosis, autophagy, and cell senescence, which were also revealed by enrichment of DEM target gene lists.
The integrated cancer pathway was also detected by microRNA target genes in both comparison pairs. In both groups, target genes involved in this pathway were identified, including AKT1, ATM, BARD1, BAX, BCL2, BRCA1, CASP3, CDK4, CHEK1, CHEK2, JAK1, MDM2, PLK1, PTEN, RB1, SMAD2, SMAD3, TP53. Analysis revealed multiple interactions between the identified microRNA target genes, indicating their significant contribution and joint involvement in tumor growth processes. Functional clustering of the target genes showed their involvement in cell cycle regulation, DNA repair signaling pathways, pathways activated by estrogens and androgens, growth factors, and interleukins. Figure 3 shows the clustering and interconnection of target genes as well as their involvement in cellular processes and intracellular signaling pathways.
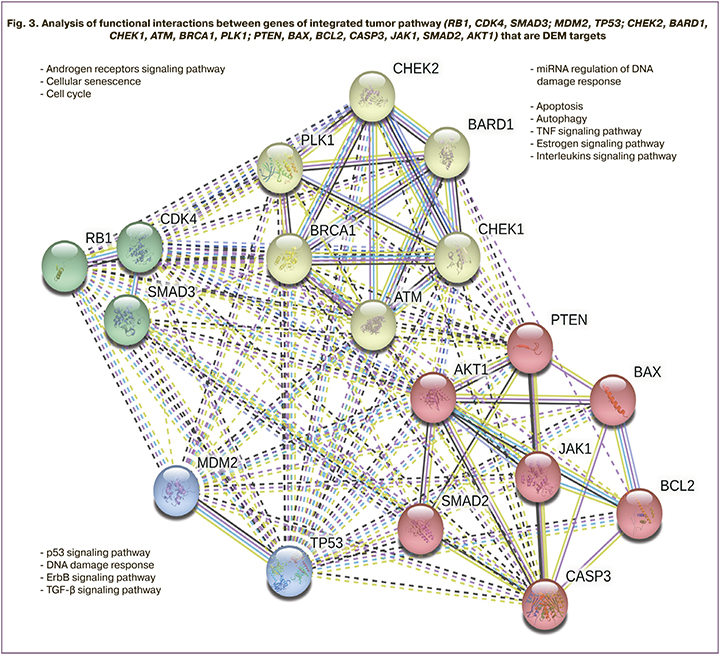
Thus, our analysis of microRNAs identified in exosomes confirms their specificity for pathogenetic processes associated with the investigated proliferative ovarian pathologies.
Discussion
The study of exosomes is a promising area of molecular medicine due to their constituent nucleic acids, proteins, glycoconjugates, lipids and even organelles - these components dynamically reflect pathophysiological conditions [17], participate in intercellular communication [10, 11] and are protected from external enzymatic effects by lipid bilayer of vesicular membrane [18]. In the present study, transcriptome analysis of circulating exosomal plasma microRNA expression was performed in patients with stage III–IV serous ovarian cancer and patients with disseminated extragenital endometriosis and OEC was performed. The list of microRNAs was shown to be altered in the blood of patients with ovarian cancer and OEC, compared to blood samples from the control groups.
The number of reads in sequencing is a parameter of the relative representation of microRNAs in the study sample. Low number of reads significantly reduce the probability of detecting and using candidate microRNAs in a comparative analysis. Therefore, DEMs with the highest possible number of reads and multiplicity of changes should be selected as potential markers. Based on these criteria, 11 microRNAs were selected (Table 2).
Changes in microRNA representation may be due not only to a specific disease, but also to hormonal changes characteristic of menstrual cycle phases [19, 20] and periods of reproductive system activity [21] in patients of different ages. However, the data remain contradictory. In a study by S. Moustafa et al., the determination of a list of microRNAs (miR-125b-5p, miR-150-5p, miR-342-3p, miR-451a, miR-3613-5p, let-7b) allowed a highly accurate differentiation of patient samples with mild to severe endometriosis (AUC=0.94) regardless of menstrual cycle phase and hormonal drug intake (combined oral contraceptives, progesterone, etc.) [22]. N. Zafari et al. also reported no effect of menstrual cycle phase on miR-199b-3p and miR-224-5p expression, whereas changes of let-7d-3p in patients with endometriosis were correlated with cycle phase, while those in the observed control group were not [20]. To test the hypothesis of a confounding effect of age, we compared microRNA expression in healthy volunteers of reproductive and postmenopausal age. In spite of the fact that statistically significant differences in the expression of 26 exosomal presumably "age-associated" microRNAs were obtained, comparison of their expression in healthy volunteers and patients with proliferative processes revealed no coincidence, which testifies to the absence of age factor influence on the representation of these microRNAs in ovarian cancer and OEC. This means that the use of DEM data as markers will not lead to false positive results. On the one hand, this makes the use of the proposed microRNAs (Table 2) relevant for the detection of ovarian cancer and OEC in different age groups and, in particular, for their use as markers of difficulties in the differential diagnosis of these proliferative processes in advanced stages. However, on the other hand, taking into account the limitations of the pilot study, we believe that the age factor should still be taken into account in the potential use of microRNAs.
Our findings for some markers were compared with the results of published studies and are shown in Table 4.
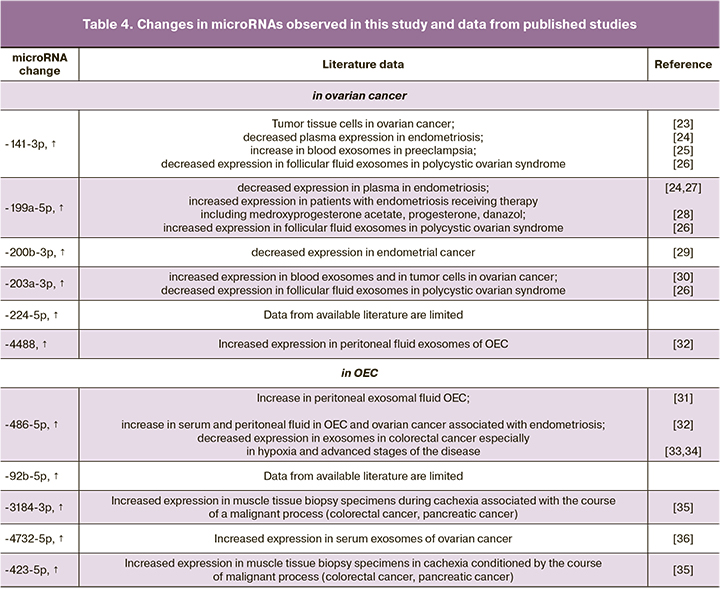
According to the literature, miRNAs of the miR-200, let-7 and miR-199 families are the most studied in ovarian diseases [19, 20, 37]. Q. Cao et al. [38] showed the prognostic value of miRNA determination in tissues of high-grade serous ovarian cancer. The increase of miR-200a, miR-200b, and miR-200c expression is associated with decreased overall survival. We found increased expression of members of this family (miR-203a-3p, miR-200b-5p and miR-141-3p), which can be interpreted as a representation of processes occurring in tumor tissue. The advantage is the detection of changes using a noninvasive method. The role of this marker in the survival of patients with advanced stages of ovarian cancer may also be studied in the future. Regarding the detected increase in miR-141-3p in ovarian cancer, S. Masoumi-Dehghi et al. showed that miR-141-3p induces endothelial growth factor receptor type 2 (VEGFR-2) activity, which intensifies tumor angiogenesis [23]. While studying the activity of JAK-STAT3 signaling pathway in ovarian cancer cell cultures, the authors showed that tumor-secreted exosomal miR-141-3p microRNAs, acting as important mediators of intercellular communication, suppress cytokine-inducible suppressors (SOCS5) activity, which results in JAK-STAT3 (The Janus kinase/signal transducer and activator of transcription) activity stimulation in endothelial cells. Our findings showed increased expression of miR-203a-3p (padj=0.011) in advanced stages of serous ovarian cancer. Similar findings were reported by D. Taylor et al. in 2008 [30]. The authors also showed similar changes in blood plasma exosomes and directly in tumor cells. These changes in the expression of this microRNA can be explained by epigenetic regulation, including hypomethylation.
We selected miR-486-5p as a marker of advanced endometriosis with OEC (padj=0.025), which confirms the reproducibility of the microRNA study. N. Nakamura et al. showed increased serum and peritoneal fluid expression of this microRNA in ovarian cancer compared to changes in endometriosis. Due to the dependence of miR-486-5p expression on the degree of disease spread, the determination of this microRNA was proposed as a marker of endometriosis severity. They also demonstrated the role of this microRNA in the induction of proliferation in OEC and ovarian cancer associated with OEC [32].
In addition to the comparable data, we obtained results that were not previously published in the diseases studied (Table 4) or differing from the previously published data. Previously, J. Liu et al. showed that when differentiating blood samples of patients with ovarian cancer from those of control patients, the determination of miR-4732-5p in plasma was characterized by 85.7% sensitivity and 82.4% specificity (AUC=0.889, p<0.0001) [36]. The authors also showed that this microRNA is an informative marker for monitoring patients with early stages to detect disease progression (p=0.018). Unlike the above study, we found no statistically significant change in miR-4732-5p in patients with serous ovarian cancer, compared to patients with OEC and control subjects. It should be noted that we observed changes in this microRNA in OEC patients who had an increase in its expression compared to control subjects (padj=0.015).
For a more in-depth study of the biological role of the identified DEMs, we analyzed the regulatory function of potential 1288 targets in several signaling pathways most likely involved in the pathogenesis and differentiation of the studied proliferative processes. On the basis of the most statistically significant differences identified, target genes of various signaling pathways were identified. They included AKT1, ATM, BARD1, BAX, BCL2, BRCA1, CASP3, CDK4, CHEK1, CHEK2, JAK1, MDM2, PLK1, PTEN, RB1, SMAD2, SMAD3, TP53 (Table 2). Functional clustering of these target genes showed their involvement in the processes of cell cycle regulation, growth factor signaling pathways, apoptosis, and DNA repair, which may confirm their specificity for pathogenic processes associated with the studied proliferative ovarian pathologies. The similarity of the analyzed groups in the activation of several signaling pathways was found. There were numerous interactions between the identified microRNA target genes that indicate their significant contribution and joint involvement in the processes of proliferative activity in ovarian cancer and OEC. The regulation mechanisms are described in detail in several studies, in particular, in the 2021 review by Gajek et al. [39], as well as in the original studies [40, 41].
Conclusion
Due to the stability of blood plasma exosomes, the study of its constituent microRNAs is a promising area of search for markers of proliferative processes. The presence of DEM in blood plasma exosomes is indicative of specific changes in ovarian cancer and OEC. The following DEMs were chosen as markers for ovarian cancer and OEC: miR-141-3p -199a-5p -200b-3p, -203a-3p, -224-5p, -4488 (increased in ovarian cancer), miR-486-5p, -3184-3p, -4732-5p, -423-5p, -92b-5p (increased in OEC).
The possible involvement in the signaling pathways of the exosomal microRNAs selected in the first stage of the study allowed to identify them as candidates for markers due to their specificity. Creating a classifying cluster confirms the feasibility of using liquid biopsy to investigate the marker potential of exosomal microRNAs.
References
1. Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017; 67(1):7-30. https://dx.doi.org/10.3322/caac.21387.2017; 67(1): 7-30.
2. Serdar E. Bulun M.D. Endometriosis. N. Engl. J. Med. 2009; 360: 268-79. https://dx.doi.org/10.1056/NEJMra0804690.
3. Павлович С.В., Юрова М.В., Мелкумян А.Г., Франкевич В.Е., Хабас Г.Н., Чаговец В.В. Биомаркеры при новообразованиях яичников: возможности, ограничения и перспективы применения у женщин репродуктивного возраста. Акушерство и гинекология. 2019; 11: 65-73. [Pavlovich S.V., Yurova M.V., Melkumyan A.G., Frankevich V.E., Chagovets V.V., Khabas G.N. Biomarkers in ovarian neoplasms: opportunities, limitations, and prospects for using in reproductive-aged women. Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2019; 11: 65-73. (in Russian)]. https://dx.doi.org/10.18565/ aig.2019.11.65-73.
4. Fassbender A., Burney R.O., O D.F., D'Hooghe T., Giudice L. Update on biomarkers for the detection of endometriosis. Biomed. Res. Int. 2015; 2015: 130854. https://dx.doi.org/10.1155/2015/130854.
5. Aznaurova Y.B., Zhumataev M.B., Roberts T.K., Aliper A.M., Zhavoronkov A.A. Molecular aspects of development and regulation of endometriosis. Reprod. Biol. Endocrinol. 2014 Jun 13; 12:50. https://dx.doi.org/10.1186/ 1477-7827-12-50.
6. Cortez M.A., Bueso-Ramos C., Ferdin J., Lopez-Berestein G., Sood A.K., Calin G.A. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011; 8(8): 467-77. https://dx.doi.org/10.1038/nrclinonc.2011.76.
7. Mirzaei H., Gholamin S., Shahidsales S., Sahebkar A., Jaafari M.R., Mirzaei H.R. et al. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur. J. Cancer. 2016; 53: 25-32. https://dx.doi.org/10.1016/ j.ejca.2015.10.009.
8. Iorio M.V., Visone R., Di Leva G., Donati V., Petrocca F., Casalini P. et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007; 67(18): 8699-707. https://dx.doi.org/10.1158/0008-5472.CAN-07-1936.
9. Filigheddu N., Gregnanin I., Porporato P.E., Surico D., Perego B., Galli L. et al. Differential expression of micrornas between eutopic and ectopic endometrium in ovarian endometriosis. J. Biomed. Biotechnol. 2010; 2010: 369549. https://dx.doi.org/10.1155/2010/369549.
10. Anfossi S., Babayan A., Pantel K., Calin G.A. Clinical utility of circulating non-coding RNAs — an update. Nat. Rev. Clin. Oncol. 2018; 15(9): 541-63. https://dx.doi.org/10.1038/s41571-018-0035-x.
11. Farooqi A.A., Desai N.N., Qureshi M.Z., Librelotto D.R.N., Gasparri M.L., Bishayee A. et al. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol. Adv. 2018; 36(1): 328-34. https://dx.doi.org/10.1016/j.biotechadv.2017.12.010.
12. Giannopoulou L., Zavridou M., Kasimir-Bauer S., Lianidou E.S. Liquid biopsy in ovarian cancer: the potential of circulating miRNAs and exosomes. Transl. Res. 2019; 205: 77-91. https://dx.doi.org/10.1016/j.trsl.2018.10.003.
13. Aboutalebi H., Bahrami A., Soleimani A., Saeedi N., Rahmani F., Khazaei M. et al. The diagnostic, prognostic and therapeutic potential of circulating microRNAs in ovarian cancer. Int. J. Biochem. Cell Biol. 2020; 124: 105765. https://dx.doi.org/10.1016/j.biocel.2020.105765.
14. Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15(12): 550. https://dx.doi.org/10.1186/s13059-014-0550-8.
15. Licursi V., Conte F., Fiscon G., Paci P. MIENTURNET: An interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinformatics. 2019; 20(1):545. https://dx.doi.org/10.1186/ s12859-019-3105-x.
16. Glaab E., Baudot A., Krasnogor N., Schneider R., Valencia A. EnrichNet: network-based gene set enrichment analysis. Bioinformatics. 2012; 28(18): i451- 7. https://dx.doi.org/10.1093/bioinformatics/bts389.
17. Pegtel D.M., Gould S.J. Exosomes. Annu. Rev. Biochem. 2019; 88: 487-514. https://dx.doi.org/10.1146/annurev-biochem-013118-111902.
18. Kim K.M., Abdelmohsen K., Mustapic M., Kapogiannis D., Gorospe M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA. 2018; 8(4): 10.1002/ wrna.1413. https://dx.doi.org/10.1002/wrna.1413.
19. Cho S., Mutlu L., Grechukhina O., Taylor H.C. Circulating microRNAs as potential biomarkers for endometriosis. Fertil. Steril. 2015; 103(5): 1252-60. e1. 103(5):1252- 103(5):1252-60. e1. https://dx.doi.org/10.1016/ j.fertnstert.2015.02.013.
20. Zafari N., Tarafdari A.M., Izadi P. A panel of plasma miRNAs 199b-3p, 224-5p and Let-7d-3p as non-invasive diagnostic biomarkers for endometriosis. Reprod. Sci. 2021; 28(4): 991-9. https://dx.doi.org/10.1007/ s43032-020-00415-z.
21. Vanhie A., O D., Peterse D., Beckers A., Cuellar A., Fassbender A. et al. Plasma miRNAs as biomarkers for endometriosis. Hum. Reprod. 2019; 34(9): 1650-60. https://dx.doi.org/10.1093/humrep/dez116.
22. Moustafa S., Burn M., Mamillapalli R., Nematian S., Flores V., Taylor H.S. Accurate diagnosis of endometriosis using serum microRNAs. Am. J. Obstet. Gynecol. 2020; 223(4): 557.e1-557.e11. https://dx.doi.org/10.1016/ j. ajog. 2020.02.050.
23. Masoumi-Dehghi S., Babashah S. microRNA-141-3p-containing small extracellular vesicles derived from epithelial ovarian cancer cells promote endothelial cell angiogenesis through activating the JAK / STAT3 and NF-к B signaling pathways. J. Cell Commun. Signal. 2020; 14(2): 233-44. https://dx.doi.org/10.1007/s12079-020-00548-5.
24. Leonova A., Turpin V.E., Agarwal S.K., Leonardi M., Foster W.G. A critical appraisal of the circulating levels of differentially expressed microRNA in endometriosis. Biol. Reprod. 2021; 105(5): 1075-85. https://dx.doi.org/10.1093/ biolre/ioab134.
25. Truong G., Guanzon D., Kinhal V., Elfeky O., Lai A., Longo S. et al. Oxygen tension regulates the miRNA profile and bioactivity of exosomes released from extravillous trophoblast cells — Liquid biopsies for monitoring complications of pregnancy. PLoS One. 2017; 12(3): e0174514. https://dx.doi.org/10.1371/ journal.pone.0174514.
26. Hu J., Tang T., Zeng Z., Wu J., Tan X., Yan J. The expression of small RNAs in exosomes of follicular fluid altered in human polycystic ovarian syndrome. PeerJ. 2020; 8: e8640. https://dx.doi.org/10.7717/peerj.8640.
27. Papari E., Noruzinia M., Kashani L., Foster W.G. Identification of candidate microRNA markers of endometriosis with the use of nextgeneration sequencing and quantitative real-time polymerase chain reaction. Fertil. Steril. 2020; 113(6): 1232-41. https://dx.doi.org/10.1016/ j.fertnstert.2020.01.026.
28. Bjorkman S., Taylor H.S. MicroRNAs in endometriosis: biological function and emerging biomarker candidates. Biol. Reprod. 2019; 100(5): 1135-46. https://dx.doi.org/10.1093/biolre/ioz014.
29. Roman-Canal B., Moiola C.P, Gatius S., Bonnin S., Ruiz-Miro M., Gonzalez E. et al. EV-associated miRNAs from peritoneal lavage are a source of biomarkers in endometrial cancer. Cancers (Basel). 2019; 11(6): 839. https://dx.doi.org/10.3390/cancers11060839.
30. Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008; 110(1): 13-21. https://dx.doi.org/10.1016/j.ygyno.2008.04.033.
31. Chen Y., Wang K., Xu Y., Guo P., Hong B., Cao Y. et al. Alteration of myeloid-derived suppressor cells, chronic inflammatory cytokines, and exosomal miRNA contribute to the peritoneal immune disorder of patients with endometriosis. Reprod. Sci. 2018; 26(8): 1130-8. https://dx.doi.org/10.1177/1933719118808923.
32. Nakamura N., Terai Y., Nunode M., Kokunai K., Konishi H., Taga S. et al. The differential expression of miRNAs between ovarian endometrioma and endometriosis-associated ovarian cancer. J. Ovarian Res. 2020; 13(1): 51. https://dx.doi.org/10.1186/s13048-020-00652-5.
33. Bj0rnetr0 T., Redalen K.R., Meltzer S., Thusy anthan N.S., Samiappan R., Jegerschold C. et al. An experimental strategy unveiling exosomal microRNAs 486-5p, 181a-5p and 30d-5p from hypoxic tumour cells as circulating indicators of high-risk rectal cancer. J. Extracell. Vesicles. 2019; 8(1): 1567219. https://dx.doi.org/10.1080/20013078.2019.1567219.
34. Liu C., Li M., Hu Y., Shi N., Yu H., Liu H., Lian H. miR-486-5p attenuates tumor growth and lymphangiogenesis by targeting neuropilin-2 in colorectal carcinoma. Onco Targets Ther. 2016; 9: 2865-71. https://dx.doi.org/10.2147/ OTT.S103460.
35. Narasimhan A., Ghosh S., Stretch C., Greiner R., Bathe O.F., Baracos V., Damaraju S. Small RNAome pro fi ling from human skeletal muscle: novel miRNAs and their targets associated with cancer cachexia. J. Cachexia Sarcopenia Muscle. 2017; 8(3): 405-16. https://dx.doi.org/10.1002/ jcsm.12168.
36. Liu J., Yoo J., Ho J.Y., Jung Y., Lee S., Hur S.Y., Choi Y.J. Plasma-derived exosomal miR-4732-5p is a promising noninvasive diagnostic biomarker for epithelial ovarian cancer. J. Ovarian Res. 2021; 14(1): 59. https://dx.doi.org/10.1186/s13048-021-00814-z.
37. Cochrane D.R., Howe E.N., Spoelstra N.S., Richer J.K. Loss of miR-200c: a marker of aggressiveness and chemoresistance in female reproductive cancers. J. Oncol. 2010; 2010: 821717. https://dx.doi.org/10.1155/2010/821717.
38. Cao Q., Lu K., Dai S., Hu Y., Fan W. Clinicopathological and prognostic implications of the miR-200 family in patients with epithelial ovarian cancer. Int. J. Clin. Exp. Pathol. 2014; 7(5): 2392-401.
39. Gajek A., Gralewska P., Marczak A., Rogalska A. Current implications of microRNAs in genome stability and stress responses of ovarian cancer. Cancers (Basel). 2021; 13(11): 2690. https://dx.doi.org/10.3390/cancers13112690.
40. Бобров М.Ю., Балашов И.С., Филиппова Е.С., Альмова И.К., Тимофеева А.В., Гусар В.А., Боровиков П.И., Хилькевич Е.Г., Чупрынин В.Д., Павлович С.В. Оценка экспрессии микроРНК в очагах ретроцервикаль- ного эндометриоза. Акушерство и гинекология. 2018; 6: 55-61. [Bobrov M.Yu., Balashov I.S., Filippova E.S., Almova I.K., Timofeeva A.V., Gusar V.A., Borovikov P.I., Khilkevich E.G., Chuprynin V.D., Pavlovich S.V. Assessment of microRNA expression in retrocervical endometriotic lesions. Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2018; 6: 55-61. (in Russian)]. https://dx.doi.org/10.18565/aig.2018.6.55-61.
41. Бобров М.Ю., Балашов И.С., Филиппова Е.С., Альмова И.К., Хилькевич Е.Г., Павлович С.В., Наумов В.А., Боровиков П.И., Сухих Г.Т. Использование транскриптомных баз данных для анализа патогенетических факторов эндометриоза. Акушерство и гинекология. 2017; 4: 34-44. [Bobrov M.Yu., Balashov I.S., Filippova E.S., Almova I.K., Khilkevich E.G., Pavlovich S.V., Naumov V.A., Borovikov P.I., Sukhikh G.T. Use of transcriptomic databases for the analysis of pathogenetic factors of endometriosis. Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2017; 4: 34-44. (in Russian)]. https://dx.doi.org/10.18565/aig.2017.4.34-44.
Received 11.01.2022
Accepted 25.02.2022
About the Authors
Mariia V. Iurova, Ph.D. Student at the Chair of Obstetrics, Gynecology, Perinatology and Reproductology, I.M. Sechenov First MSMU, Ministry of Health of Russia (Sechenov University), 119991, Russia, Moscow, Trubetskaya str., 8-2, +7(495)438-20-88; Specialist, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 117997, Russia, Moscow, Ac. Oparina str., 4, m_yurova@oparina4.ru, https://orcid.org/0000-0002-0179-7635Chupalav M. Eldarov, Ph.D. (Bio), Senior Researcher at the Laboratory of Molecular Pathophysiolog, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 117997, Moscow, Russia, Ac. Oparina str., 4, +7(495)438-77-00, ch_eldarov@oparina4.ru Mikhail Yu. Bobrov, Ph.D. (Chemistry), Head of the Laboratory of Molecular Pathophysiology, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 117997, Moscow, Russia, Ac. Oparina str., 4, mbobr@mail.ru
Grigory N. Khabas, Ph.D., surgeon, oncologist, obstetrician-gynecologist, Head of the Department of Innovative Oncology and Gynecology, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 117997, Russia, Moscow, Ac. Oparina str., 4, +7(495)438-78-33, g_khabas@oparina4.ru, https://orcid.org/0000-0002-5011-9152
Stanislav V. Pavlovich, PhD, Professor, Department of Obstetrics, Gynecology, Perinatology, and Reproductology, Faculty of Postgraduate Professional Training of Physicians, I.M. Sechenov First Moscow State Medical University, Ministry of Health of Russia (Sechenov University); Academic Secretary, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 117997, Russia, Moscow, Ac. Oparina str., 4., +7(495)438-20-88, s_pavlovich@oparina4.ru, https://orcid.org/0000-0002-1313-7079
Corresponding author: Mariia V. Iurova, +7(926)385-45-48, hi5melisa@gmail.com, m_yurova@oparina4.com
Authors' contributions: lurova M.V. - conception and design of the study, analysis of relevant literature, database creation and maintenance, statistical analysis and interpretation of clinical data, manuscript drafting and editing, preparation of documentation; Bobrov M.Yu. - design of the study, carrying out all stages of the practical part of the study, manuscript editing, biological interpretation of the data, final approval of the manuscript, preparation of documentation; Eldarov Ch.M. - design of the study, analysis of relevant literature, work with databases, statistical analysis and interpretation of data, manuscript drafting and editing, preparation of documentation; Khabas G.N. - surgical treatment of patients, clinical interpretation of the data, critical revision and editing of the manuscript, final approval of the he manuscript; Pavlovich S.V. - design of the presented study, editing, clinical interpretation of the data, final approval of the manuscript, preparation of documentation.
Conflicts of interest: The authors have no conflicts of interest to declare.
Funding: The study was conducted within the framework of the state order of Ministry of Health of Russia "Development of approaches for noninvasive diagnostics of endometriosis based on omics technologies"; state registration number: АААА-А19-119021490132-9 for the period 01.01.19-31.12.21.
Ethical Approval: The study was reviewed and approved by the Research Ethics Committee of the I.M. Sechenov First MSMU, Ministry of Health of Russia (Sechenov University) (Ref. No. 16-19 of 05.12.2019) and the Research Ethics Committee of the V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia (Ref. No. 10 of 05.12.2019).
Patient Consent for Publication: All patients provided informed consent for the publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator.
For citation: Iurova M.V., Eldarov Ch.M., Bobrov M.Yu., Khabas G.N., Pavlovich S.V. Expression of exosomal microRNA in high-grade ovarian cancer and ovarian endometriotic cysts.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2022; 3: 68-79 (in Russian)
https://dx.doi.org/10.18565/aig.2022.3.68-79



