Noninvasive diagnosis of embryo chromosomal status in vitro: integration of raman spectrometry and machine learning in assisted reproductive technologies
Valiakhmetovaa E.Z., Rimskaya E.N., Gorevoy A.V., Yakimova A.S., Sysoeva A.P., Ekimov A.N., Makarova N.P., Kalinina E.A., Sukhikh G.T.
Relevance: This study is relevant due to the relatively low efficiency of in vitro fertilization (IVF), which has an implantation success rate of up to 40%, as well as the invasiveness of current preimplantation genetic testing (PGT-A) used to detect chromosomal abnormalities in embryos. The invasive nature of these procedures poses a risk of embryo damage, highlighting the need for innovative and noninvasive approaches.
Objective: To develop a noninvasive method for assessing the chromosomal status of embryos using spectral analysis of spent culture media and machine learning techniques.
Materials and methods: The study involved 36 couples, from whom 40 samples of spent culture media were obtained (11 from euploid embryos and 29 from aneuploid embryos). Raman scattering (RS) spectra were recorded using a Confotec MR520 microscope-spectrometer with 532 nm laser excitation. Machine learning algorithms, including quadratic discriminant analysis (QDA), combined with stratified fivefold cross-validation, were employed to analyze the spectral characteristics and differentiate between the sample groups.
Results: Significant differences were observed in the mean RS spectra of spent culture media between euploid and aneuploid embryos. The most reliable discriminating features included the intensity ratios of Raman bands at 735 cm-1 (phosphatidylserine, DNA), 1196 cm-1 (nucleic acids), and 1666 cm-1 (C=C stretching, amide I), respectively. The developed predictive model achieved an accuracy, sensitivity, and specificity of 84 %, 80%, and 88%, respectively.
Conclusion: Raman spectra of embryo culture media obtained with 532 nm laser excitation may reveal novel biochemical indicators associated with embryonic developmental abnormalities. These findings provide new perspectives for noninvasive diagnostics in reproductive medicine and have the potential to enhance the effectiveness of assisted reproductive technology programs.
Authors' contributions: Valiakhmetova E.Z., Makarova N.P., Rimskaya E.N., Gorevoy A.V. – conception and design of the study; Sysoeva A.P., Yakimova A.S. – collection and processing of material; Makarova N.P., Valiakhmetova E.Z. – statistical analysis; Valiakhmetova E.Z., Makarova N.P., Rimskaya E.N. – drafting of the manuscript; Kalinina E.A., Sukhikh G.T. – editing of the manuscript.
Conflicts of interest: The authors have no conflicts of interest to declare.
Funding: The study was conducted as part of the initiative research project “Study of the influence of extracellular vesicles of biological fluids of reproductive organs and tissues on gametes, the process of fertilization and early human embryogenesis and implantation” (2025–2027, supervisor Makarova N.P.). V.I. Kulakov NMRC for OG&P, Ministry of Health of Russia.
Ethical Approval: The study was reviewed and approved by the Research Ethics Committee of the V.I. Kulakov NMRC for OG&P.
Patient Consent for Publication: All patients provided informed consent for the publication of their data.
Authors' Data Sharing Statement: The data supporting the findings of this study are available upon request from the corresponding author after approval from the principal investigator.
For citation: Valiakhmetovaa E.Z., Rimskaya E.N., Gorevoy A.V., Yakimova A.S., Sysoeva A.P., Ekimov A.N., Makarova N.P., Kalinina E.A., Sukhikh G.T. Noninvasive diagnosis of embryo chromosomal status in vitro:
integration of raman spectrometry and machine learning in assisted reproductive technologies.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2026; (1): 78-88 (in Russian)
https://dx.doi.org/10.18565/aig.2025.251
Keywords
Infertility remains one of the most pressing medical and social problems in reproductive health, affecting approximately 15–20% of married couples of reproductive age [1, 2]. Assisted reproductive technologies (ART), particularly in vitro fertilization (IVF), are the primary means of addressing reproductive disorders; however, the effectiveness of these programs is relatively low, with successful implantation rates not exceeding 40% [2].
A critical factor influencing IVF outcomes is embryo quality, which is directly related to the chromosomal status. Aneuploidies, a type of chromosomal abnormality, are the leading cause of implantation failure, early reproductive loss, and infertility. Existing methods of preimplantation genetic testing (PGT), such as next-generation sequencing (NGS), are invasive, require trophectoderm biopsy, and carry a certain risk of embryo damage. [3].
The need for noninvasive, highly accurate methods to assess the genetic potential of embryos has driven the search for novel diagnostic approaches. A promising direction is the use of Raman spectrometry, a highly sensitive method for analyzing biological fluids that provides molecular composition data without damaging biological samples [4, 5]. Of particular relevance is the integration of machine learning methods into embryo selection for transfer during IVF [6, 7].
This study aimed to develop a noninvasive method for assessing the chromosomal status of embryos during infertility treatment using ART, based on spectral analysis of spent culture media combined with machine learning.
Materials and methods
Patient enrollment and description
The study was conducted at Prof. B.V. Leonov Department for Assisted Reproductive Technologies, V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology, and Perinatology, Ministry of Health of Russia. Spectral characterization of spent culture media and implementation of machine learning algorithms were performed at the Clinical Proteomics Laboratory of the same center and at the Laboratory of Laser Nanophysics and Biomedicine, Lebedev Physics Institute, Russian Academy of Sciences.
Patients were enrolled after providing written informed consent and meeting specific inclusion criteria: preserved ovarian reserve, regular menstrual cycles, retrieval of at least one embryo of good or excellent morphological quality according to the Gardner classification during an IVF or intracytoplasmic sperm injection (ICSI) cycle, and indication for preimplantation genetic testing for aneuploidy (PGT-A) due to advanced reproductive age, severe male-factor infertility, or repeated IVF failure (≥ 3 previous unsuccessful attempts). The exclusion criteria included general contraindications defined in Order No. 803n of the Ministry of Health of the Russian Federation (July 31, 2020) "On the procedure for the use of assisted reproductive technologies, contraindications, and limitations", as well as the use of donor oocytes, premature ovarian insufficiency, endometrial pathology, interstitial and/or subserosal uterine myoma >4 cm, or submucosal myoma deforming the uterine cavity.
A total of 36 couples were included, five of whom used donor sperm for IVF/ICSI treatment. Embryos assessed as good or excellent underwent PGT-A using next-generation sequencing (NGS) at the Laboratory of Preimplantation Genetic Testing and Genetic Diagnostics, Institute of Reproductive Genetics, V.I. Kulakov NMRC OG&P. All couples underwent pre-cycle evaluation in accordance with Order No. 803n of the Ministry of Health of the Russian Federation and the 2024 clinical guidelines "Female Infertility."
Ovarian stimulation and embryology procedures
Controlled ovarian stimulation was initiated on days 2–3 of the menstrual cycle using a standard GnRH antagonist protocol with highly purified human menopausal gonadotropin and/or recombinant gonadotropins. The initial dose was individualized based on patient characteristics (body mass index, age, ovarian reserve, and previous stimulation history) and adjusted according to the dynamics of follicular and endometrial growth. To prevent premature luteinizing hormone (LH) surge, a GnRH antagonist was administered subcutaneously at 0.25 mg/day once the leading follicle reached 13–14 mm in diameter. When the dominant follicle reached ≥17 mm, ovulation was triggered 35 h before transvaginal oocyte retrieval using either human chorionic gonadotropin (5,000–10,000 IU, intramuscularly) or recombinant chorionic gonadotropin alpha (250 μg, subcutaneously). In 11 patients, a GnRH agonist trigger (0.1–0.2 mL, subcutaneously) was used to minimize the risk of ovarian hyperstimulation syndrome. Oocyte retrieval was performed 35 h after triggering under intravenous anesthesia and transvaginal ultrasound guidance in aseptic conditions. Subsequent oocyte assessment and fertilization were performed according to standard laboratory procedures. All embryological procedures were conducted in GLOBAL culture media (CooperSurgical, Denmark) according to the manufacturer's instructions. Given the requirement for subsequent PGT-A, fertilization was performed using ICSI in all cases. Embryos were cultured under low-oxygen conditions (5%) until day 5 without medium renewal in individual droplets under a layer of mineral oil (Vitrolife, Sweden). On day 5, samples of spent culture media and trophectoderm biopsies were collected for PGT-A. The embryos were cryopreserved by vitrification (Protein Synthesis, Russia).
Trophectoderm Biopsy and PGT-A
On day 5 after fertilization, trophectoderm biopsy was performed on blastocysts of good or excellent morphological quality (grades 3-6 AB, BA, AA) using borosilicate biopsy needles (Origio, Denmark). The cells were transferred into Eppendorf tubes containing lysis buffer and sent to the molecular genetics laboratory for analysis. Embryos were cultured for 1-2 hours after biopsy and vitrified if no prominent degeneration was observed.
Collection of spent culture media for Raman profiling
Samples of spent culture medium (25 μL each) were collected aseptically from under the mineral oil layer into Eppendorf tubes and stored at -20°C until analysis. A total of 40 samples were obtained; according to PGT-A results, 11 corresponded to chromosomally normal (euploid) embryos and 29 to aneuploid embryos.
Raman spectroscopy
For spectral acquisition, the samples were thawed at room temperature (25°C). A 1 μL droplet of each sample was placed in a liquid form on an aluminum oxide-coated glass slide. Measurements were taken using a Confotec MR520 confocal Raman microscope (SOL Instruments, Minsk, Belarus) excited by a 532 nm laser (20 mW at the sample plane, 5 s accumulation time) and a 40× MPlanFL objective (NA 0.75; Nikon, Japan). The spectral range was 400-1900 cm⁻¹ (fingerprint region). According to the manufacturer specifications, the lateral and axial optical resolutions were <1 μm and <10 μm, respectively, and the spectral resolution ranged from 1 to 1.5 cm⁻¹. Between 5 and 10 spectra were collected from random points for each sample under identical conditions.
Machine learning-based spectral analysis
Raman spectra were pre-processed using implementations of the Vancouver Raman algorithm [4, 8, 9] in MATLAB (R2022b, MathWorks, Massachusetts, USA), applying modified multipolynomial baseline fitting for fluorescence removal and Savitzky-Golay smoothing for noise reduction. Each spectrum was approximated using a set of pseudo-Voigt functions fitted using the least-squares method to determine the peak positions corresponding to significant Raman bands. Hidden peaks were identified using the second derivative method [10].
To assess the diagnostic potential of spectral features for distinguishing culture media from euploid and aneuploid embryos, several classifiers from the MATLAB Classification Learner were tested, including linear discriminant analysis (LDA) and quadratic discriminant analysis (QDA). Unlike decision trees or k-nearest neighbors (kNN), QDA enables the derivation of complete receiver operating characteristic (ROC) curves and estimation of specificity for a given sensitivity (and vice versa), providing a more interpretable metric of classification performance.
We applied stratified five-fold cross-validation repeated ten times and calculated the mean ± standard deviation of the classification metrics (e.g., specificity) for each model. Stratification ensured the proportional representation of both outcome classes (euploid/aneuploid) in the training and test sets, consistent with the full dataset. Data splitting was performed at the sample level, not at the individual spectrum level [4], to prevent an overly optimistic bias arising from nearly identical spectra from the same sample being present in both the training and test subsets.
Results
In the first stage, we evaluated the clinical and anamnestic data of the enrolled patients (Table 1). The mean age of the women was 38 years, and their body mass indices were within the normal range. The menstrual function of the participants exhibited typical reproductive patterns. The mean age at menarche was 13 years, which is consistent with the normative indicators of physiological development. The average menstrual cycle length was 28 days, with minor variations between 27.5 and 30 days, which aligns with physiological norms. The mean duration of menstruation was 5 days, and the mean age at sexual debut was 20 years, indicating a socially mature approach to initiating sexual activity. Menstrual irregularities were reported by only 8.3% of the participants, suggesting no significant reproductive dysfunction in the study group. Overall, the data indicate a favorable background for menstrual function, with minimal deviations from physiological norms.
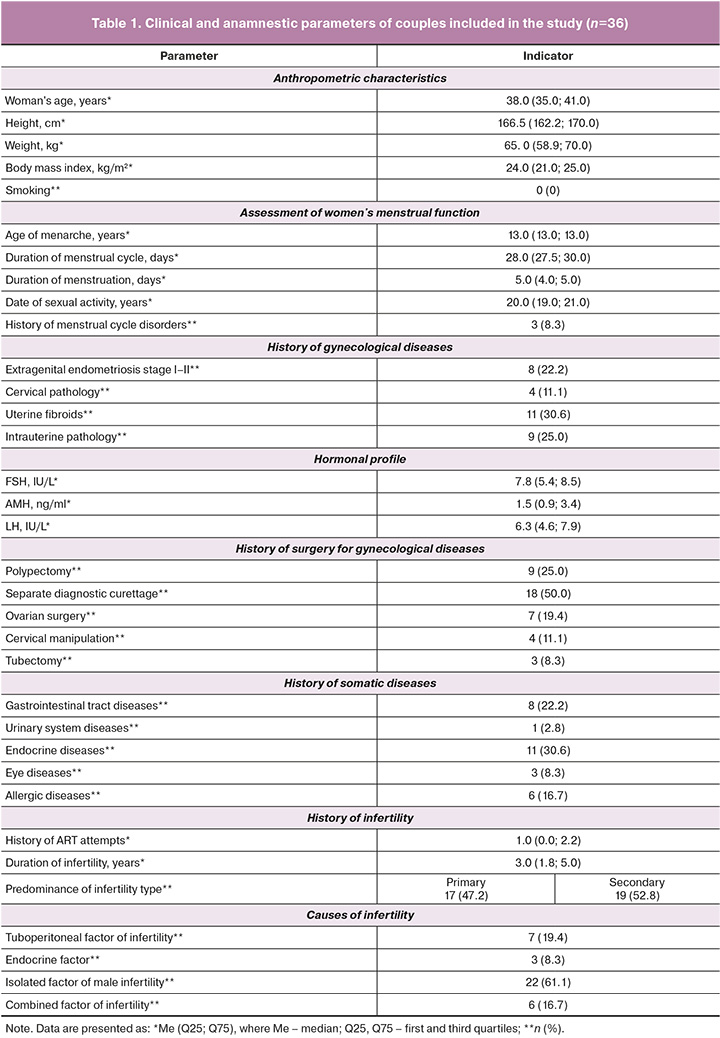
Hormonal assessments revealed that the average follicle-stimulating hormone (FSH) level was 7.8 IU/L, which was within the reference range for reproductive-age women. The mean luteinizing hormone (LH) level was 6.3 IU/L, which was also within the normal limits. The anti-Müllerian hormone (AMH) concentration was 1.5 ng/mL, indicating a moderate decrease in ovarian reserve, consistent with the expectations for women aged 38 years. Uterine fibroids were the most common gynecological condition, diagnosed in 30.6% of women, indicating their notable prevalence. Intrauterine pathology was found in 25.0% of cases, and stage I-II external genital endometriosis was identified in 22.2% of patients. Cervical pathology was recorded in 11.1% of the cases. These findings suggest a significant presence of reproductive abnormalities among women in the study group who underwent preimplantation genetic testing for aneuploidy (PGT-A) as part of infertility treatment. In most cases (30.6%), somatic comorbidities were endocrine disorders that had been managed by specialists prior to ART treatment.
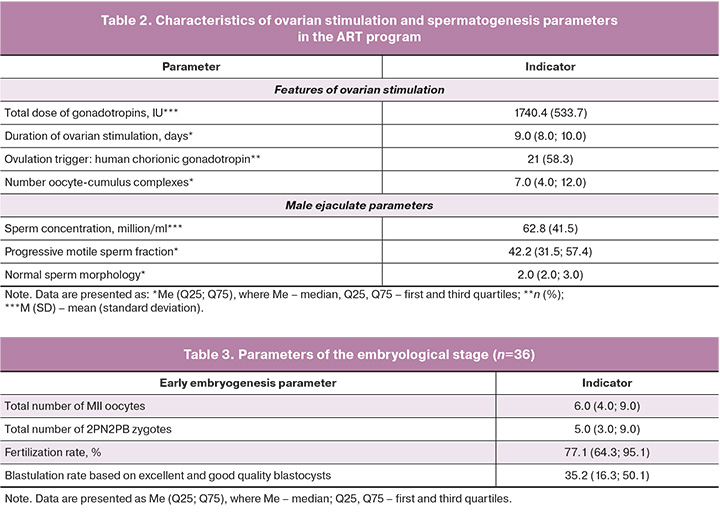
The characteristics of ovarian stimulation and spermatogenesis parameters in the ART program are summarized in Table 2. The standard ovarian stimulation protocol using a GnRH antagonist resulted in an average of six oocytes per patient, with a mean total gonadotropin dose of 1740.4 IU administered. Semen parameters on the day of fertilization indicated teratozoospermia, with a mean sperm concentration of 62.8 million/mL and 42.2% of progressively motile sperm. The embryological phase observed in the enrolled couples showed typical outcomes for women aged >35 years: the mean fertilization rate was 77.1%, and the proportion of excellent- and good-quality blastocysts was 35.2% (Table 3).
Subsequently, we performed spectroscopy of the spent culture medium and developed a prognostic model to estimate the probability of determining an embryo's genetic status based on its metabolic activity, using machine learning algorithms.
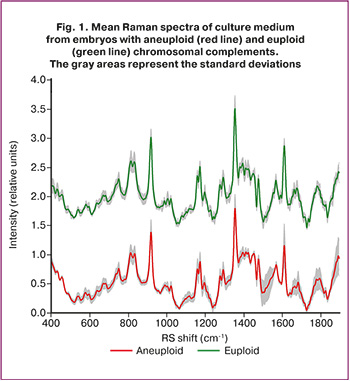
Figure 1 shows the mean Raman spectra (RS) of 40 samples of spent culture medium obtained from embryos with normal (euploid) and abnormal (aneuploid) chromosomal constitutions, as identified by PGT-A.
The normalized mean spectra (colored curves) with standard deviation intervals (gray shading), as illustrated in Figure 1, demonstrate the stability of the spectral characteristics. Greater variability in spectral intensities within specific bands indicates higher diversity in the intrinsic properties of samples from different patients and their corresponding spectral distributions. For euploid RS spectra, pronounced deviations were observed at approximately 1050–1150 cm⁻¹, 1490–1580 cm⁻¹, 1650–1700 cm⁻¹, and approximately 1800 cm⁻¹, while for aneuploid spectra, deviations were noted at approximately 830 and 1380–1450 cm⁻¹.
RS spectra were obtained for 11 euploid embryos (90 spectra) and 29 aneuploid embryos (220 spectra) using laser excitation at 532 nm across a range of 400–2000 cm⁻¹. The main Raman band positions of the culture medium derived from the embryos are shown in Figure 2. The processed RS spectra in the range of 400–2000 cm⁻¹ for all culture medium samples showed similar major RS bands but differed in peak positions and intensities. We began our analysis by identifying distinct spectral peaks and investigating their potential correspondence with the key components of the culture medium.
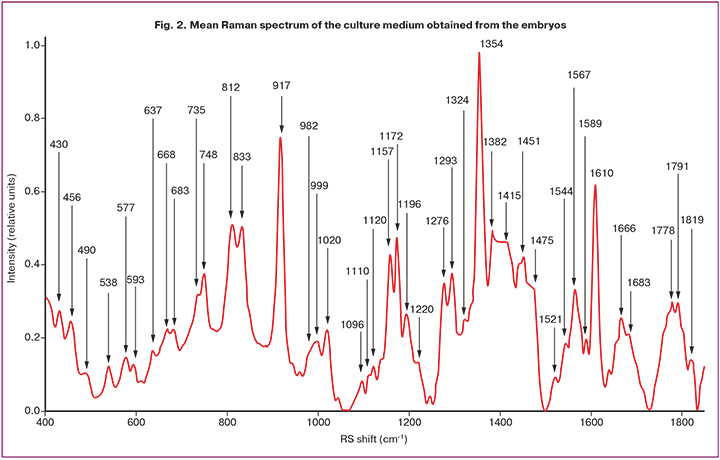
Significant differences between the RS spectra of euploid and aneuploid embryos allowed their differentiation using Raman light spectroscopy. Consequently, we developed a method to identify abnormal embryos based on spectral criteria related to the ratios of biomarker-specific Raman bands in the spent culture medium. Analyzing the most representative RS bands that provide the highest classification scores is valuable for elucidating biochemical changes during the development of abnormalities, where the relative content of proteins, lipids, and nucleic acids (DNA/RNA) may serve as potential biomarkers. The analysis of 42 bands at an excitation wavelength of 532 nm revealed distinct differences in their relative intensities between aneuploid and euploid embryos.
For a detailed analysis, each spectrum was represented as an array of its peak intensities across all observed RS bands, as shown in Figure 2. To determine the most effective criteria for differentiation, each band was sequentially used as a reference; the intensities of the other bands were divided by it, and all possible intensity ratio pairs were considered. Receiver operating characteristic (ROC) curves were plotted for each pair using a quadratic discriminant analysis (QDA) classifier, with the pair providing the highest specificity at 80% sensitivity. Our tests indicated that the most reliable classification results were obtained when normalization was performed on the mean spectral intensity.
Comparing the RS spectra of euploid and aneuploid embryos revealed that the main differences were primarily in peak intensities, indicating altered concentrations of specific compounds. Notably, the ratios between the peaks at 735, 1196, and 1666 cm⁻¹ differed substantially. The RS band at 735 cm⁻¹ typically corresponds to phosphatidylserine, although it may also reflect changes in the DNA. The band at 1196 cm⁻¹ is attributed to nucleic acid vibrations of adenine, guanine, and cytosine. The band at 1666 cm⁻¹ corresponds to the C=C stretching vibrations (amide I). The distributions of the three most informative spectral features are shown in Figure 3.
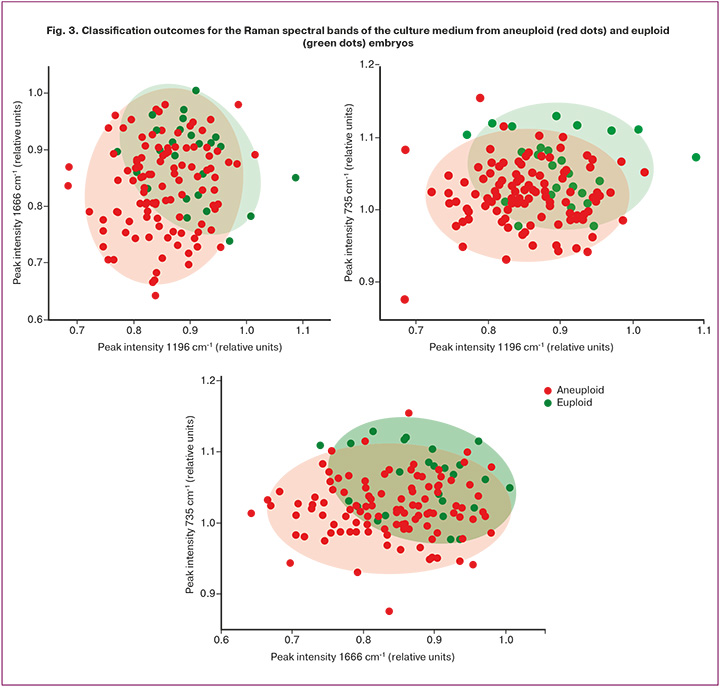
The differentiation of aneuploid and euploid embryos at an excitation wavelength of 532 nm demonstrated that the bands at 735, 1196, and 1666 cm⁻¹ provided the highest classification performance, achieving 84% accuracy, 80% sensitivity, and 88% specificity.
Discussion
Metabolic stress induced by aneuploidy is a complex phenomenon that arises from the disruption of the delicate balance of metabolic pathways within the cell. Under normal conditions, embryonic physiology critically depends on the precise coordination of these pathways and the stoichiometrically balanced abundance of enzymes and regulatory factors. Aneuploidy alters gene dosage, disrupting this balance and leading to significant metabolic changes in the cell. Our data demonstrate that it is possible to noninvasively and accurately predict the genetic status of an embryo based on the identified differences in the metabolic profiles of euploid and aneuploid embryos. Raman spectrometry is particularly well-suited for predicting ploidy, as it enables an integrated assessment of metabolic stress reflected in the composition of the culture medium. Differences in metabolic processes between embryos with normal and abnormal genotypes have been previously demonstrated [14]. Cytogenetic abnormalities associated with chromosomal defects lead to proteotoxic, metabolic, replicative, and mitotic stress. Consequently, abnormal genotypes generate metabolic profiles distinct from those of genetically normal embryos. This observation is supported by studies analyzing culture media samples from embryos with known PGT results [15].
The findings of the present study align with the conclusions of several scientific publications on metabolomic profiling of embryo culture media, which aim to identify associations between detected signatures and embryo quality (including genetic quality) as well as transfer outcomes [15, 16]. In a study by Liang et al., three Raman spectral ranges were identified that enabled the classification of embryo genetic status with an accuracy of up to 95.9% [15]. The three most significant biochemical markers (ranges 967–1015 cm⁻¹, 1129–1295 cm⁻¹, and 1400–1430 cm⁻¹), in combination with machine learning, facilitated the development of a predictive model, followed by data validation. According to the authors, metabolic markers within these ranges correspond to small noncoding RNAs (981 cm⁻¹), lipids (1420 cm⁻¹), and U/C ring stretching vibrations (1249 cm⁻¹). In our study, one of the three significant markers may have originated from lipids. Another study described differences in amino acid metabolism between euploid and aneuploid embryos, demonstrating that tyrosine concentrations in the culture media of aneuploid embryos were significantly higher than those in euploid embryos (76.38 μmol/L, p<0.003) [17]. Spent culture media from genetically normal and abnormal embryos on days 2–3 of cultivation differed in the intensity of asparagine, glycine, and valine exchange, while on days 3–4, they exhibited differences in serine, leucine, and lysine concentrations. Differences have also been observed in pentose phosphate pathway enrichment, vitamin B6 metabolism, and amino acid metabolism in embryos with different karyotypes [18].
In our study, the metabolic markers identified for differentiating the genetic status of embryos were associated with phosphatidylserine (735 cm⁻¹), vibrations of nitrogenous bases (guanine, adenine, cytosine; 1196 cm⁻¹), and C=C valence vibrations (amide I; 1666 cm⁻¹).
The synthesis of nitrogenous bases during early embryonic development involves the pentose phosphate pathway. According to the scientific data cited above, there is a significant correlation between embryo karyotype and enrichment of the pentose phosphate pathway, which, together with our findings, provides an integrated view of the metabolic transformations occurring in embryos with normal and abnormal karyotypes. In studies of mammalian embryogenesis, nitrogenous base synthesis has been shown to regulate the activation of trophectoderm gene transcription, which is potentially associated with impaired implantation and placentation processes [19].
Phosphatidylserine in early embryogenesis is linked to apoptosis, differentiation, and tissue maturation [20].
In the present study, we present the results of Raman scattering (RS) spectroscopic analysis of 40 spent culture media samples from embryos with known genetic statuses. The feasibility of differentiating euploid from aneuploid embryos based on the RS spectra of the culture medium was confirmed. A machine learning algorithm for ploidy prediction, achieving an accuracy of 84%, was developed, and specific spectral biomarkers correlated with embryo chromosomal status were identified.
Conclusion
The intensity ratios of the bands at 735, 1196, and 1666 cm⁻¹ were the most reliable criteria for three-class differentiation under 532 nm excitation. Thus, acquiring RS spectra of embryo culture media using 532 nm excitation may reveal novel biochemical indicators of abnormal embryonic development in the future. The results obtained open new prospects for noninvasive diagnostics in reproductive medicine with the potential to enhance the effectiveness of ART programs.
References
- Blockeel C., Campbell A., Coticchio G., Garcia-Velasco J.A., Pinborg A., Santulli P. Educate. Empower. Reproduce. A call for action against the demographic winter. Reprod. Biomed. Online. 2025; 51(4): 105047. https://dx.doi.org/10.1016/j.rbmo.2025.105047
- Назаренко Т.А., ред. Бесплодный брак. Клинические задачи и их решение. М.: МЕДпресс-информ; 2024. 144 с. [Nazarenko T.A., ed. Infertile marriage. Clinical problems and their solutions. Moscow: MEDpress-Inform; 2024. 144 p. (in Russian)].
- Лисицына О.И., Романов А.Ю., Сыркашева А.Г., Макарова Н.П., Долгушина Н.В. Влияние биопсии трофэктодермы на течение беременности и акушерские исходы. Проблемы репродукции. 2025; 31(3): 63-9. [Lisitsyna O.I., Romanov A.Yu., Syrkasheva A.G., Makarova N.P., Dolgushina N.V. Impact of trophectoderm biopsy on the course of pregnancy and obstetric outcomes. Russian Journal of Human Reproduction. 2025; 31(3): 63-9 (in Russian)]. https://dx.doi.org/10.17116/repro20253103163
- Rimskaya E., Gorevoy A., Yakimova A., Makarova N., Starodubtseva N., Kudryashov S. et al. Enhancing male fertility diagnostics with seminal plasma Raman spectroscopy. Spectrochim Acta. A. Mol. Biomol. Spectrosc. 2025; 340: 126237. https://dx.doi.org/10.1016/j.saa.2025.126237
- Zakaria A., Diawara I., Bouziyane A., Louanjli N. Exploring human sperm metabolism and male infertility: a systematic review of genomics, proteomics, metabolomics, and imaging techniques. Int. J. Mol. Sci. 2025; 26(15): 7544. https://dx.doi.org/10.3390/ijms26157544
- Драпкина Ю.С., Макарова Н.П., Чаговец В.В., Васильев Р.А., Амелин В.В., Калинина Е.А. Использование машинного обучения для анализа липидного профиля среды культивирования и прогнозирования эффективности вспомогательных репродуктивных технологий. Акушерство и гинекология. 2025; 2: 91-9. [Drapkina Yu.S., Makarova N.P., Chagovets V.V., Vasiliev R.A., Amelin V.V., Kalinina E.A. Using machine learning to analyze the lipid profile of culture medium and predict the efficacy of assisted reproductive technologies. Obstetrics and Gynecology. 2025; (2): 91-9 (in Russian)]. https://dx.doi.org/10.18565/aig.2024.280
- Драпкина Ю.С., Макарова Н.П., Васильев Р.А., Амелин В.В., Калинина Е.А. Сравнение прогностических моделей, построенных с помощью разных методов машинного обучения, на примере прогнозирования результатов лечения бесплодия методом вспомогательных репродуктивных технологий. Акушерство и гинекология. 2024; 2: 97-105. [Drapkina Yu.S., Makarova N.P., Vasiliev R.A., Amelin V.V., Kalinina E.A. Comparison of predictive models built with different machine learning techniques using the example of predicting the outcome of assisted reproductive technologies. Obstetrics and gynecology. 2024; 2: 97-105 (in Russian)]. https://dx.doi.org/10.18565/aig.2023.263
- Rimskaya E., Gorevoy A., Shelygina S., Perevedentseva E., Timurzieva A., Saraeva I. et al. Multi-wavelength raman differentiation of malignant skin neoplasms. Int. J. Mol. Sci. 2024; 25(13): 7422. https://dx.doi.org/10.3390/ijms25137422
- Zhao J., Lui H., McLean D.I., Zeng H. Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy. Appl. Spectrosc. 2007; 61(11): 1225-32. https://dx.doi.org/10.1366/000370207782597003
- Saraeva I.N., Rimskaya E.N., Timurzieva A.B., Gorevoy A.V., Sheligyna S.N., Popadyuk V.I. et al. Analysis of skin neoplasms’ Raman spectra using the Lorentz approximation method: pilot studies. JETP Lett. 2024; 119(7): 556-63. https://dx.doi.org/10.1134/S0021364023604153
- Rival C.M., Xu W., Shankman L.S., Morioka S., Arandjelovic S., Lee C.S. et al. Phosphatidylserine on viable sperm and phagocytic machinery in oocytes regulate mammalian fertilization. Nat. Commun. 2019; 10(1): 4456. https://dx.doi.org/10.1038/s41467-019-12406-z
- Böse J., Gruber A.D., Helming L., Schiebe S., Wegener I., Hafner M. et al. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 2004; 3(4): 15. https://dx.doi.org/10.1186/jbiol10
- Talari A.C.S., Movasaghi Z., Rehman S., Rehman I. Raman spectroscopy of biological tissues. Applied Spectroscopy Reviews. 2014; 50(1): 46-111. https://dx.doi.org/10.1080/05704928.2014.923902
- Zhu J., Tsai H.J., Gordon M.R., Li R. Cellular stress associated with aneuploidy. Dev. Cell. 2018; 44(4): 420-31. https://dx.doi.org/10.1016/j.devcel.2018.02.002
- Liang B., Gao Y., Xu J., Song Y., Xuan L., Shi T. et al. Raman profiling of embryo culture medium to identify aneuploid and euploid embryos. Fertil. Steril. 2019; 111(4): 753-62.e1. https://dx.doi.org/10.1016/j.fertnstert.2018.11.036
- Sánchez-Ribas I., Riqueros M., Vime P., Puchades-Carrasco L., Jönsson T., Pineda-Lucena A. et al. Differential metabolic profiling of non-pure trisomy 21 human preimplantation embryos. Fertil. Steril. 2012; 98(5): 1157-64.e1-2. https://dx.doi.org/10.1016/j.fertnstert.2012.07.1145
- Olcay I.O., Akcay B., Bahceci M., Arici A., Boynukalin K., Yakicier C. et al. Noninvasive amino acid turnover predicts human embryo aneuploidy. Gynecol. Endocrinol. 2022; 38(6): 461-6. https://dx.doi.org/10.1080/09513590.2022.2068520
- Eldarov C., Gamisonia A., Chagovets V., Ibragimova L., Yarigina S., Smolnikova V. et al. LC-MS analysis revealed the significantly different metabolic profiles in spent culture media of human embryos with distinct morphology, karyotype and implantation outcomes. Int. J. Mol. Sci. 2022; 23(5): 2706. https://dx.doi.org/10.3390/ijms23052706
- Chi F., Sharpley M.S., Nagaraj R., Roy S.S., Banerjee U. Glycolysis-independent glucose metabolism distinguishes TE from ICM fate during mammalian embryogenesis. Dev. Cell. 2020; 53(1): 9-26.e4. https://dx.doi.org/10.1016/j.devcel.2020.02.015
- Brooks E.C., Zeidler M.P., Ong A.C.M., Evans I.R. Macrophage subpopulation identity in Drosophila is modulated by apoptotic cell clearance and related signalling pathways. Front. Immunol. 2024; 14: 1310117. https://dx.doi.org/10.3389/fimmu.2023.1310117
Received 15.09.2025
Accepted 29.12.2025
About the Authors
Elvira Z. Valiakhmetova, PhD student at the Prof. B.V. Leonov Department for Assisted Technologies in Infertility Treatment, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 117997, Russia, Moscow, Ac. Oparin str., 4, ibraeva1988@list.ruElena N. Rimskaya, PhD, Senior Researcher at the Laboratory of Clinical Proteomics, Academician V.I. Kulakov National Medical Research Center for Obstetrics,
Gynecology and Perinatology, Ministry of Health of Russia, 117997, Russia, Moscow, Ac. Oparin str., 4; Researcher at the Laboratory of Laser Nanophysics and Biomedicine,
Center for Laser and Nonlinear Optical Technologies, Department of Quantum Radiophysics named after N.G. Basov, P.N. Lebedev Physics Institute of RAS (FIAN),
119991, Russia, GSP-1 Moscow, Leninsky Ave., 53, rimskaya@lebedev.ru, https://orcid.org/0000-0001-7802-0720
Alexey V. Gorevoy, Researcher at the Laboratory of Laser Nanophysics and Biomedicine, Center for Laser and Nonlinear Optical Technologies, Department of Quantum Radiophysics named after N.G. Basov, P.N. Lebedev Physics Institute of RAS (FIAN), 119991, Russia, GSP-1, Moscow, Leninsky Ave., 53, a.gorevoy@lebedev.ru,
https://orcid.org/0000-0003-4208-0291
Alexandra S. Yakimova, Embryologist at the Prof. B.V. Leonov Department for Assisted Technologies in Infertility Treatment, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 117997, Russia, Moscow, Ac. Oparin str., 4, yakimoovaal@gmail.com,
https://orcid.org/0009-0001-5913-2660
Anastasia P. Sysoeva, PhD, Embryologist at the Prof. B.V. Leonov Department for Assisted Technologies in Infertility Treatment, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 117997, Russia, Moscow, Ac. Oparin str., 4, a_sysoeva@oparina4.ru,
https://orcid.org/0000-0002-6502-4498
Аlexey N. Ekimov, PhD, Head of the Laboratory of Preimplantation Genetic Testing and Genetic Diagnostics, Academician V.I. Kulakov National Medical Research Center
for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 4, Oparina str., Moscow, 117997, Russia, a_ekimov@oparina4.ru,
https://orcid.org/0000-0001-5029-0462
Natalya P. Makarova, Dr. Bio. Sci., Leading Researcher at the Prof. B.V. Leonov Department of Assistive Technologies in Infertility Treatment, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 117997, Russia, Moscow, Ac. Oparin str., 4,
np_makarova@oparina4.ru, https://orcid.org/0000-0003-1396-7272
Elena A. Kalinina, Dr. Med. Sci., Professor, Head of the Prof. B.V. Leonov Department of Assistive Technologies in Infertility Treatment, Academician V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia, 117997, Russia, Moscow, Ac. Oparin str., 4, e_kalinina@oparina4.ru, https://orcid.org/0000-0002-8922-2878
Gennady T. Sukhikh, Dr. Med. Sci., Professor, Academician of the RAS, Director, Academician V.I. Kulakov National Medical Research Center for Obstetrics,
Gynecology and Perinatology, Ministry of Health of Russia, 117997, Russia, Moscow, Ac. Oparin str., 4, g_sukhikh@oparina4.ru, https://orcid.org/0000-0002-7712-1260



