Эндометриоз – заболевание, которое характеризуется формированием эктопических очагов эндометриоидной ткани за пределами матки, имеющих разнообразную локализацию в брюшной полости: в ретроцервикальном пространстве, на ретровагинальной перегородке, мочевом пузыре, брюшине, матке, прямой кишке, аппендиксе и яичнике [1].
Формирование, развитие и распространение эктопических очагов стимулирует процессы неспецифического воспаления, сопровождающегося болевым синдромом, фиброзированием эктопической эндометриоидной ткани, приводящим к развитию спаечного процесса в брюшной полости. При множественной локализации очагов эндометриоза наблюдается вовлечение в патологический процесс сразу нескольких органов, что приводит к тяжелым осложнениям, связанным с нарушением их физиологических функций. Распространенность эндометриоза в общей популяции может достигать 10%, а среди пациенток с бесплодием и/или болевым синдромом – до 50% [2]. Важно отметить, что эффективного медикаментозного лечения данного заболевания до сих пор не предложено. На данный момент существует несколько концепций этиологии и патогенеза эндометриоза [3]. Тем не менее, ключевые механизмы развития заболевания, позволяющие выявить молекулярные мишени для терапевтического воздействия, остаются недостаточно изученными.
Одним из перспективных подходов в исследовании патогенеза эндометриоза является применение так называемых «омиксных» технологий, основанных на широкомасштабном анализе различных классов молекул в нормальных и патологически измененных клетках и тканях. Постепенное повышение доступности методов геномного, транскриптомного, протеомного и метаболомного анализа приводит к накоплению ценной информации о количественных и качественных структурно-функциональных изменениях на соответствующих уровнях молекулярной организации. При этом возникает новый класс задач, связанный с анализом получаемых массивов данных. Необходимо отметить, что применение всех перечисленных технологий в рамках одного исследования одной лабораторией на сегодняшний день представляется проблематичным. Рациональным подходом в данной ситуации является совместное использование и анализ данных, получаемых разными научными группами, применяющими альтернативные «омиксные» подходы или их комбинации. Таким образом, коллективное использование информации может позволить получать оптимальное количество данных для формирования относительно целостной картины изучаемых явлений.
В рамках данного подхода были созданы базы данных, например GEO (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo), где исследователи делятся результатами экспериментов, полученных с использованием различных омиксных технологий. Открытость этой и других баз позволяет на этапе планирования и подготовки эксперимента провести анализ полученных другими исследователями данных для формулировки гипотез и целей предстоящих исследований, а также сопоставить данные, полученные в ходе одного исследования, с результатами других авторов. Важным аспектом анализа баз данных является возможность построения путей и сетей предполагаемых молекулярных взаимодействий, характерных для определенных физиологических и патологических процессов. Интеграция биологической информации, полученной при помощи различных высокопроизводительных технологий может служить эффективным подходом для выявления ключевых механизмов развития заболеваний.
В данной работе был проведен анализ транскриптомных наборов данных из базы GEO для поиска ключевых регуляторов и мишеней в этиологии и патогенезе эндометриоза. В исследование включено шесть наборов данных (GDS2737 [4], GDS5339 [5], GDS3060 [6], GDS3975 [7], GDC3092 [8], GDS2835 [9]), найденных в соответствующих статьях в результате поиска по ключевым словам «endometriosis» и «endometrium». Использованные наборы данных были получены авторами статей при помощи транскриптомного микроматричного анализа образцов тканей человека. В зависимости от использованных групп сравнения, наборы были поделены на парные и непарные. В парных наборах авторы исследований проводили сравнение экспрессии между эктопическим и эутопическим эндометрием одного пациента. В непарных наборах проводилось сравнение ткани эктопического очага одного пациента с тканью эндометрия другого пациента, не болеющего эндометриозом. Основные характеристики наборов данных приведены в табл. 1.
Наборы данных GDS2737 и GDS5339 были разбиты на несколько подгрупп. Набор GDS5339 включал две группы парных (GDS5339_1) и непарных (GDS5339_2) образцов. Набор GDS2737 был разделен в соответствии с фазой цикла: GDS2737_1 – ранняя секреторная фаза, GDS2737_2 – средняя секреторная фаза, GDS2737_3 – пролиферативная фаза.
Нами был проведен анализ транскриптомных данных и выделены дифференциально экспрессированные гены из каждого набора. Затем на основе списка этих генов было проведено обогащение метаболических путей, молекулярных функций и биологических процессов.
Для выявления возможных механизмов регуляции экспрессии генов был также проведен анализ данных по дифференциальной экспрессии микроРНК в тканях эндометрия. Молекулы микроРНК, входящие в состав белкового комплекса RISC, ингибируют процесс трансляции за счет взаимодействия с мРНК гена-мишени. В результате такого взаимодействия происходит временная остановка трансляции, либо мРНК деградирует, и трансляции не происходит [10]. Таким образом, экспрессию микроРНК целесообразно учитывать при анализе экспрессии мРНК-транскриптов, так как данная информация может объяснить несоответствие транскриптомных данных и результатов, полученных при количественных и функциональных исследованиях соответствующих белков. В данной работе, нами было рассмотрено 9 исследований, в которых экспрессия микроРНК изучалась с помощью микрочиповой технологии [11–13], ПЦР [14–17] и с использованием высокопроизводительного секвенирования [7, 18].
Ввиду различия панелей и дизайна исследований, наборы данных включающие в себя информацию о транскриптоме, были проанализированы раздельно. Нормализация данных проводилась при помощи встроенных средств сервиса GEO, анализ дифференциальной экспрессии проводился при помощи Orange Canvas 3.38 [19]. В качестве дифференциально экспрессированных рассматривались гены, со значением |FC|>2 (|FC| – модуль кратности изменения средних значений экспрессии в группах), скорректированному по методу FDR с p-value <0.05. Обогащение (enrichment) путей внутриклеточной сигнализации и биологических процессов по базе данных KEGG проведено при помощи программы Cytoscape 3.4.09 [20] с использованием плагина JEPETTO10 [21]. Поиск таргетных мРНК для микроРНК проводилась при помощи базы валидированных взаимодействий miRWalk 2.0 11 [22].
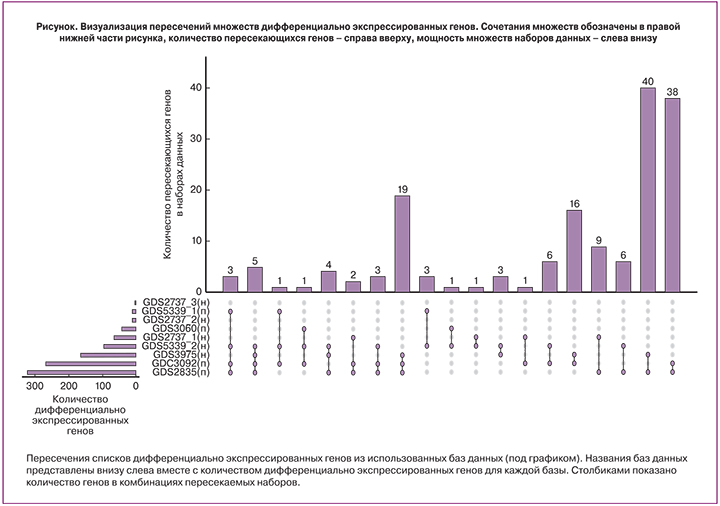
В ходе исследования в использованных транскриптомных наборах было обнаружено 1019 с повышенной и пониженной экспрессией в эндометриозных очагах. При пересечении списков дифференциально экспрессированных генов, обнаружено 168 генов, встречающихся более чем в 1 наборе. При этом для наборов с парными образцами число таких генов составило 86, а для наборов с непарными образцами – 14. Следует отметить совпадение направленности изменения экспрессии среди всех оцениваемых генов в разных наборах своей группы (парных и непарных). Ряд транскриптов (3 в группе непарных наборов, 18 в группе парных), не имеющих функциональной аннотации, был исключен из дальнейшего анализа (рисунок).
Список генов, полученных при пересечении отобранных баз данных можно характеризовать низкой чувствительностью к влиянию особенностей исследований и используемых методических платформ, то есть выбранные гены демонстрировали дифференциальную экспрессию и одинаковую направленность изменений вне зависимости от принадлежности к тому или иному набору. Необходимо подчеркнуть, что в большинстве исследований проводится оценка экспрессии генов в тканях эндометрия, которые состоят из различных типов клеток, по-разному представленных в нормальной и патологически измененной ткани. Поэтому при оценке вклада экспрессии того или иного гена нужно учитывать тот факт, что мы имеем суммарный результат всех типов клеток в изучаемом образце.

В результате анализа наборов парных и непарных данных по выбранным параметрам, были получены списки дифференциально экспрессирующихся генов в эктопическом эндометрии относительно эутопического эндометрия. В наборе парных данных было отобрано 70 генов, а в непарных 9 генов, четыре из которых определялись также в парном наборе. В исследование были включены аннотированные гены, для которых идентифицированы кодируемые ими белки и описано их биологическое значение. На основе функциональной роли белков, кодируемых выявленными генами, было проведено разбиение на функциональные группы: белки межклеточного матрикса, рецепторы, мембранные белки, белки цитоскелета, ферменты и ингибиторы, белки внутриклеточных каскадов (киназы, фосфатазы, GTP-азы, адаптеры), транскрипционные факторы и ДНК-ассоциированные белки. Списки генов для каждой функциональной группы парных и непарных образцов приведены в табл. 2. Для функциональной характеристики дифференциально экспрессированных генов использовались данные литературных источников, а также аннотации приведенные в базе данных Gene Национального центра биотехнологической информации США (NCBI, http://www.ncbi.nlm.nih.gov). После аббревиатуры гена будет приведено обозначение направленности его экспрессии (+ или -) в очагах эндометриоза относительно эутопического эндометрия.
Внеклеточный матрикс
В группу белков внеклеточного матрикса распределился ген белка ECM1 (-), который по некоторым данным принимает участие в регуляции клеточной миграции, инвазии и адгезии. В линиях опухолевых клеток, снижение экспрессии ECM1 приводило к перестройкам цитоскелета и снижению экспрессии прометастатических регуляторов TGFβR2 и S100A4, последний также был идентифицирован в парном наборе данных (см. табл. 2) [23].
Рецепторы
Среди генов рецепторов (9 представителей: 6 с повышенной «+» и 3 с пониженной «-» экспрессией) можно выделить: FZD7 (+) – рецептор участвующий в запуске Wnt-сигнального пути [24], GPR1 (+) – рецептор, связывающий хемотаксический пептид химерин, и запускающий ERK1/2 каскад [25], RXFP1(-) – рецептор для релаксинов, активация которого приводит к повышению синтеза сАМР и активации РКА, запуску ERK1/2 пути, повышению синтеза NO [26], SORCS2 (+) – сортиллин-подобный рецептор тип 2, мутации которого ассоциированы с лейомиомой матки [27], 2FCGR3B (+) – рецептор для Fc фрагмента IgG, THRB (+) – рецептор тиреоидного гормона [28], ESR1 (-) – эстрогеновый рецептор, TRIL (+) – коактиватор TLR4 рецептора, участвующего в процессах острого и хронического воспаления [29].
Мембранные белки
Данная группа генов также представлена девятью представителями, семь из которых имеют пониженный уровень экспрессии. GPC3 или glypican 3 (+) – мембранный белок гепарансульфат-содержащий протеогликан, участвует в регуляции процессов деления и роста клеток [30], KIAA1324 (-) или ген, индуцируемый эстрогенами (EIG121), – трансмембранный белок, онкосупрессор, в частности при раке эндометрия [31]. GJB6 (-) – коннексин, входящий в состав щелевых контактов, и CDH1 (-) – кадгерин 1-го типа (Е-кадгерин, белок клеточной адгезии), который может принимать участие в распространении очагов эндометриоза [32]. PERP (-) – мембранный белок, экспрессия которого запускается онкосупрессором р53 и инициирует апоптоз за счет активации каспаз, снижение экспрессии ассоциировано с метастатическим фенотипом опухолевых клеток [33], STXBP6 (-) или amysin – участвует в процессе экзоцитоза (негативный регулятор), в частности взаимодействует с синтаксинами, образующими SNARE-комплекс, необходимый для выделения везикул [34]; также повышение экспрессии STXBP6 приводит к снижению пролиферации и миграции и стимулирует апоптоз в линиях опухолевых клеток [35], снижение экспрессии гена STX19 (-) или синтаксина 19, возможно коррелирует со снижением экспрессии STXBP6, однако их взаимодействие пока не описано.
Белки, ассоциированные с цитоскелетом
По результатам анализа было выявлено шесть генов, 4 с повышенной и 2 с пониженной экспрессией. KIF14 (+) – митотический кинезин, принимающий участие в на стадии цитокинезе митоза [36], кроме того показана способность KIF14 взаимодействовать с киназой AKT, что коррелирует с пролиферативной активностью и пониженной чувствительностью опухолевых клеток к цитостатикам [37]. HJURP (+) – участвует в сборке центромерного комплекса хромосом при митозе, а также его повышенная экспрессия замедляет клеточное старение с вовлечением р53-зависимого регуляторного пути [38].
NUF2 (-) – компонент кинетохорного комплекса хромосом, способный контролировать клеточную пролиферацию за счет регуляции циклин-зависимых киназ [39]. PDLIM3 (+) – участвует в организации цитоскелета, и может выступать в роли онкосупрессора находящегося под контролем транскрипционного активатора р53 [40]. HOOK1 (-) – участвует в ассоциации клеточных везикул с микротрубочками и сортировке внутри клетки [41], а также способен подавлять эпителиально-мезенхимальный переход клеток опухолей [42]. MYH11 (+) – тяжелая цепь миозина 11 гладких мышц, снижение экспрессии данного гена в клетках миомы матки приводило к снижению пролиферации гладкомышечных клеток и снижению синтеза коллагена 1, фибронектина и инсулиноподобного фактора роста 1 (ILGF1) [43]. Повышенная экспрессия MYTH1 может способствовать развитию фиброза.
Ферменты и ингибиторы
В данной группе представлено восемь генов с повышенной и шесть генов с пониженной экспрессией. GALNT4 (-) – ген кодирует фермент N-ацетигалактозаминилтрансферазу 4 типа, который участвует в О-гликозилировании специфических сайтов белков-мишеней. Снижение экспрессии GALNT4 ассоциировано с развитием злокачественного фенотипа опухолевых клеток за счет стимуляции их роста, миграции и инвазии, развития устойчивости к аноикису (Anoikis – одна из форм апоптоза, развивающаяся у адгезивных клеток после открепления от межклеточного матрикса) [44].
HS3ST3A1 (+) – гепарансульфат глюкозаминил 3-О-сульфотрансфераза участвует в биосинтезе протеогликанов плазматической мембраны и межклеточного матрикса, имеющих широкий спектр биологических функций (участвуют в эмбриональном развитии, клеточной адгезии, метастазировании, коагуляции крови, ангиогенезе) [45]. Было установлено увеличение количества сульфатированных гликозаминогликанов в очагах эндометриоза [46], что согласуется с повышением экспрессии HS3ST3A1. HSD11B1 (+) – гидроксистероид (11-бета) дегидрогеназа 1 катализирует превращение неактивного кортизона в кортизол – биоактивный глюкокортикоид периферических тканей. Повышенная экспрессия этого фермента была обнаружена в очагах эндометриоза [47]. MOGAT1 (-) – моноацилглицерол О-ацилтрансфераза, принимает участие в синтезе диацилглицеролов и фосфолипидов. PRSS8 (-) – ассоциированная с мембраной сериновая протеаза простазин, может выступать в роли онкосупрессора в опухолевых клетках, снижая пролиферацию и рост, за счет ингибирования Sphk1/S1P/Stat3/Akt пути [48]. Данный фермент прикрепляется к мембране за счет фосфатидилинозитола, чей биосинтез может быть снижен за счет уменьшения экспрессии MOGAT1. PTGIS (+) – синтаза простагландина I2, монооксигеназа катализирующая превращение PGH2 в PGI2 или простациклин, обладающий вазоделатирующим и антиагрегантным действием. PGI2 может стимулировать клеточную пролиферацию за счет активации PPARδ и усиления экспрессии ангиогенного ростового фактора VEGF [49]. CYP2J2 (-) – представитель семейства цитохромов Р450, участвует в образовании эпоксидов полиненасыщенных жирных кислот, которые обладают вазоактивными свойствами, регулируют процессы тканевого воспаления и выживания клеток [50]. Причем направленность регуляторного действия зависит от исходных субстратов CYP2J2: w-6 или w-3 полиненасыщенных жирных кислот.
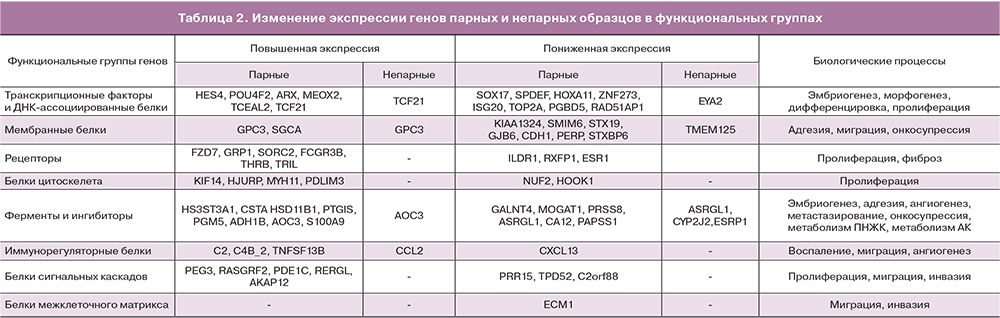
ASRGL1 (-) – L-аспарагиназа, сниженная экспрессия этого фермента, ассоциирована с неблагоприятным прогнозом при карциномах эндометрия [51].
PGM5 (+) – фосфоглюкомутаза 5, участвует в интерконверсии глюкозо-1-фосфата и глюкозо-6-фосфата, которые могут служить промежуточными метаболитами для реакций энергетического обмена, а также реакций гликозилирования [52]. ADH1B (+) – представитель семейства алкогольдегидрогеназ, участвующих в метаболизме этанола, алифатических спиртов, ретинола, гидроксистероидов и гидроперекисей липидов. AOC3 (+) – мембранный фермент, который осуществляет окислительное дезаминирование первичных аминов с образованием соответствующих альдегидов, ионов аммония и перикиси водорода, также обладает моноаминоксидазной активностью. Принимает участие в процессах воспаления и фиброзирования тканей [53]. CA12 (-) – карбоангидраза-12, мембранный фермент катализирующий гидрирование углекислого газа, недостаток может приводить к локальным сдвигам рН, в зависимости от содержания CO2 в ткани [54]. PAPSS1 (-) – фермент, обеспечивающий образование аденозин 3’-фосфат-5’-фосфосульфата, который является кофактором в реакциях сульфатирования, в частности эстрогенов, что приводит к их инактивации в тканях. Увеличение времени жизни эстрогенов в тканях за счет снижения экспрессии PAPSS1 может приводить к нарушению регуляции процессов пролиферации и апоптоза, что наблюдается, например, у гормон-чувствительных опухолей молочной железы и эндометрия [55].
CSTA (+) – ген кодирует цистатин А, ингибитор цистениновых протеаз, который регулирует расщепление белков цитозоля и цитоскелета соответствущими ферментами, а также участвует в поддержании целостности белков клеточной адгезии [56]. S100A9 (+) – ингибитор казеинкиназ, вовлечен в процессы регуляции клеточного цикла и дифференцировки, его экспрессия ассоциирована с развитием неоплазий шейки матки [57].
Белки сигнальных каскадов
В данной группе насчитывается семь генов: 4 с пониженной и 3 с повышенной экспрессией. Два гена (C2orf88 и RERGL ) пока не имеют функциональной аннотации. RASGRF2 (+) – данный ген кодирует кальций-зависимый фактор обмена гуаниловыми нуклеотидами (GDP на GTP), активирующий RAS-подобные белки, который координирует сигнальные пути митоген-активируемых киназ. Также RASGRF2 может ингибировать киназу cdc42, снижая, таким образом, подвижность и инвазивность опухолевых клеток [58]. TPD52 (-) – повышенная экспрессия данного гена отмечается в ряде опухолей различного происхождения, и коррелирует со степенью злокачественности [59], однако функции и регуляторные механизмы продуктов TPD52 остаются невыясненными. PRR15 (-) функция данного гена до конца не изучена, но по некоторым данным, его повышенная экспрессия наблюдается в непролиферирующих эпителиальных клетках при остановке клеточного цикла [60]. PDE1C (+) – фосфодиэстераза циклических нуклеотид монофосфатов (cAMP и cGMP), которые участвуют в разнообразных путях внутриклеточной сигнализации. Фермент активируется при участии ионов кальция и кальмодулина. Увеличение экспрессии PDE1C приводит к стимуляции пролиферации, миграции и инвазивности опухолевых клеток, что было продемонстрировано на линиях глиобластомы [61].
AKAP12 (+) – представитель семейства адаптерных белков, которые обеспечивают образование специфических комплексов сигнальных белков (рецепторов, каналов, киназ, фосфатаз, малых ГТФаз) в различных микродоменах клетки, в частности для регуляции клеточного цикла. Снижение экспрессии AKAP12 связывают с усилением пролиферативной активности клеток [62].
Транскрипционные факторы
SOX17 (-) – транскрипционный регулятор из семейства транскрипционных факторов SOX, участвующих в эмбриональном развитии и дифференцировке. Было установлено, что SOX17 может принимать участие в формировании желез эндометрия, возможно за счет негативной регуляции экспрессии генов, находящихся под контролем Wnt-сигнального каскада [63]. SPDEF (-) – транскрипционный фактор из семейства ETS, который также участвует в формировании желез эпителиальных выстилок воздухоносных путей и желудка за счет стимуляции дифференцировки бокаловидных клеток [64]. Интересно, что активация SPDEF стимулирует экспрессию SOX17, что может свидетельствовать о взаимосвязи понижения экспрессии SDEF и SOX17, выявленного нами при анализе баз данных. HES4 (+) – транскрипционный фактор, который также участвует в дифференцировке клеток в процессе эмбрионального развития, и является мишенью NOTCH-сигнального пути. Повышенная экспрессия HES4 коррелирует с увеличением злокачественности опухолевых клеток [65]. POU4F2 (+) – принимает участие в процессах дифференцировки клеток сетчатки, и регуляции пролиферации опухолевых клеток и их устойчивости к химиотерапии. Экспрессия POU4F2 активируется факторами роста через МАРК-киназный путь, а также эстрогенами через ESR1-эстрогеновый рецептор [66].
ARX (+) – экспрессируется в эмбриогенезе и принимает участие в развитии нервной системы; важно отметить, что одним из механизмов действия ARX является запуск Wnt-каскада передачи сигнала [67]. HOXA11 (-) – ген вовлечен в развитие матки и необходим для поддержания рецептивности эндометрия. Экспрессия HOX11 регулируется (повышается) эстрогенами, также было показано снижение экспрессии HOX11 при эндометриозе за счет метилирования промотора данного гена [68, 69]. MEOX2 (+) – может принимать участие в миогенезе при развитии конечностей позвоночных, а также было показано его участие в остановке клеточного цикла и старении эндотелиальных клеток. МEOX2 снижает активность циклин-зависимых киназ за счет активации экспрессии их ингибиторов р21 и р16 [70]. TCF21 (+) – транскрипционный фактор специфичный для тканей мезодермального происхождения. Недавно было показано, что TCF21 может взаимодействовать с эстрогеновыми рецепторами и снижать их транскрипционную активность, при этом данные эффекты зависели от SUMO-модификаций TCF21 [71]. ESRP1 (-) – регулятор сплайсинга, координирующий экспрессию эпителиальной и мезенхимальной форм рецептора фактора роста фибробластов. При снижении активности ESRP1 образуется мезенхимальная форма, таким образом, за счет альтернативного сплайсинга может регулироваться эпителиально-мезенхимальный переход [72].
Иммунорегуляторные белки
CXCL13 (-) – хемокин, участвует в реакциях тканевого иммунитета слизистых оболочек за счет рекрутирования иммунокомпетентных клеток, синтезируется эпителиальными клетками эндометрия и, возможно, играет роль в имплантации эмбриона. Было установлено, что его экспрессия зависит от фазы цикла и меняется при эндометриозе [73]. ССL2 (+) – хемокин, хемоаттрактант для моноцитов и базофилов, активно экспрессируется при патологиях, характеризующихся моноцитарными инфильтратами. Было показано, что CCL2 повышает жизнеспособность, пролиферативную активность и инвазивность культивируемых стромальных клеток эндометрия за счет активации Akt и MAPK/Erk1/2 сигнальных путей [74].
TNFSF13B (+) – цитокин, обеспечивающий активацию В-лимфоцитов. Его экспрессия была ранее показана в очагах эндометриоза [9]. С2 (+) и С4В_2 (+) – гены, кодирующие различные компоненты комплемента, могут участвовать в процессах местного воспаления. Экспрессия данных генов при эндометриозе не описана. ISG20 (-) – экзонуклеаза, стимулируемая интерфероном, участвует в реакциях врожденного иммунитета, осуществляя противовирусную защиту. Имеются данные о том, что снижение активности ISG20 приводит к ингибированию ангиогенеза [75].
Анализ процессов, регулируемых дифференциально экспрессирующимися генами, позволил выявить ряд клеточных функций и путей внутриклеточной сигнализации, нарушение которых может приводить к развитию эндометриоза. Было выявлено 56 путей, которые были разделены по вовлеченности в определенные процессы на следующие группы: пролиферация, дифференцировка, эмбриогенез, морфогенез, ангиогенез, инвазия, миграция, метаболизм, воспаление (табл. 2). Важно отметить, что для большинства выявленных генов, в научной литературе отсутствуют данные о взаимосвязи кодируемых ими белков с патогенезом эндометриоза. Большая часть исследований по экспрессии и функциональной активности выявленных генов посвящена изучению механизмов патогенеза опухолевого процесса. Однако, с учетом неопластического характера процессов развития эндометриоза, мы, с известной осторожностью, можем экстраполировать эти данные для выявления закономерностей в анализируемых результатах.
Максимально представленным по количеству задействованных генов (25) является пролиферативный процесс. С учетом комплексной и многоуровневой регуляции такого фундаментального процесса, как клеточное деление, представляется закономерным выявление при проведенном анализе групп генов, кодирующих белки клеточной адгезии, рецепторы и их лиганды, белки сигнальных каскадов и цитоскелета, ферменты и транскрипционные факторы. Также нужно отметить, что для большинства генов данной группы показано их участие в процессах клеточной миграции и инвазии.
Ткань эндометрия, с учетом ее физиологической роли, является периодически обновляющейся, что подразумевает строгий контроль над ее пролиферативной активностью. Развитие эндометриоза во многом связано с нарушением регуляции клеточного деления. Среди выявленных в очагах эндометриоза генов, пять осуществляют негативную регуляцию, а оставшиеся оказывают положительное влияние на пролиферативный процесс, что может свидетельствовать о сдвиге баланса регуляторных влияний в сторону пролиферирующего фенотипа.
Была также выявлена группа генов, участвующих в процессах тканевого воспаления. Наблюдалось изменение экспрессии генов хемокинов, цитокинов и компонентов комплимента. Кроме этого, обнаруживались гены, связанные с окислительным метаболизмом полиненасыщенных жирных кислот (ПНЖК). Оксилипины играют важную роль в регуляции иммунных реакций, в частности миграции клеток иммунной системы, продукции провоспалительных цитокинов, что подтверждается повышением экспрессии синтазы простациклина (PGI2), способного стимулировать пролиферацию, и снижением экспрессии эпоксигеназы CYP2J2, продукты которой (эпоксиды ПНЖК) способны оказывать противовоспалительное действие. Еще одним геном, задействованным в регуляции воспалительных реакций является HSD11B1, участвующий в образовании тканевого кортизола. Экспрессия данного гена снижена в эктопическом эндометрии, что может вносить свой вклад в развитие воспаления, поскольку глюкокортикоиды оказывают противовоспалительное действие.
В качестве отдельной группы можно выделить гены, участвующие в гормональном контроле функционирования эндометрия, в частности эстрогенами. Помимо снижения экспрессии гена ESR1, кодирующего эстрагеновый рецептор ERa, наблюдалось закономерное снижение экспрессии гена KIAA1324, регулируемого эстрогенами, и оказывающего онкосупрессивное действие. Транскрипционный фактор HOXA11 также стимулируется эстрогенами через ERa. Экспрессия HOXA11 была снижена в эктопическом эндометрии, что также коррелирует со снижением экспрессии гена ESR1. При этом отмечается повышение экспрессии транскрипционного фактора TCF21, который снижает транскрипционную активность эстрогеновых рецепторов. Обращает на себя внимание изменение экспрессии генов, регулирующих метаболизм эстрогенов в тканях. Так снижение экспрессии гена PAPSS1, участвующего в процессах сульфатирования эстрогенов, может приводить к снижению уровня инактивации этих гормонов. Повышение концентрации тканевых эстрогенов на фоне пониженной экспрессии ERa, может приводить к избыточной активации других подтипов эстрогеновых рецепторов и смещению баланса запускаемых ими регуляторных каскадов. С изменением концентрации тканевых эстрогенов может быть связано усиление экспрессии гена алкогольдегидрогеназы ADH1B, которая может участвовать в неспецифических превращениях гидроксистероидов. Приведенные данные показывают, что изменение эстрогеновой регуляции при эндометриозе осуществляется на уровне рецепторов и их лигандов, а также за счет влияния на транскрипционную активность лиганд-рецепторного комплекса и активность эстроген-зависимых транскрипционных факторов.
Среди проанализированных генов было выявлено 4 представителя, участвующих на разных уровнях в Wnt-сигнальном каскаде. Известно, что активация данного сигнального пути играет важную роль в таких неопластических процессах, как пролиферация, миграция и инвазия клеток [76]. Запуск каскада начинается с активации Wnt-лигандами специфических (Frizzled) рецепторов, кодируемых семейством FZD-генов. Анализ данных показал повышенную экспрессию гена Wnt-рецептора FZD7 в очагах эктопического эндометрия, а также гена транскрипционного фактора ARX, активирующего Wnt-каскад. При этом наблюдалось снижение экспрессии гена SOX17, который является негативным регулятором Wnt-каскада, и гена транскрипционного фактора SPDEF, стимулирующего экспрессию SOX17. Регуляция работы транскрипционных факторов может свидетельствовать об участии данного пути в различных аспектах жизнедеятельности клеток эндометрия. Так, имеются данные о том, что эпителиально-мезенхимальный переход, способствующий распространению эктопического эндометрия, может находиться под контролем Wnt-зависимых путей. В пользу участия Wnt-каскада в патогенезе эндометриоза свидетельствуют результаты исследований, в которых была показана повышенная секреция Wnt-лигандов стромальными клетками эктопических очагов, а также усиление синтеза коллагена при активации Wnt-каскада [77]. Таким образом, активация данного сигнального пути может участвовать как в формировании эндометриоидного очага, так и в развитии спаечного процесса. Значимым для выявления особенностей патогенеза эндометриоза представляется изменение экспрессии генов, участвующих в клеточной адгезии (GPC3, GJB6, CDH1), в эпителиально-мезенхимальном переходе (НООК1, ESPR1) и процессах фиброзирования тканей (MYTH, ADH13). Существует ли взаимосвязь между экспрессией этих генов и Wnt путем, или эти процессы идут независимо, пока не известно.
Для оценки дифференциальной экспрессии микроРНК в тканях эндометрия по данным литературы нами было рассмотрено 9 исследований, в которых экспрессия микроРНК изучалась с помощью микрочиповой технологии [11–13], ПЦР [14–17] и с использованием высокопроизводительного секвенирования [7, 18]. По данным этих исследований в тканях эндометриозных очагов наблюдается снижение (miR-202-3p, miR-424-5p, miR-449-3p, miR-556-3p, miR-126, miR-15, miR-17-5p, miR-20a, miR-200a, miR-200a, miR-200b, miR-200c, miR-182, miR-141) и повышение(miR-143, miR-145, miR-449a, miR-34c, miR-200a, miR-200b, miR-141, miR-20a, miR-29c, miR-21, miR-125a, miR-222, miR-202, miR-99a, miR-126, miR-451) экспрессии ряда микроРНК.
Увеличение экспрессии микроРНК может приводить как к снижению уровня мРНК-транскриптов за счет их деградации, так и к повышению в случае, если синтез мРНК продолжается, но при этом трансляция заблокирована. При снижении экспрессии микроРНК можно ожидать увеличение экспрессии мРНК-мишеней. Нами был проведен поиск мишеней дифференциально экспрессированных микроРНК среди описанных выше генов. Было установлено, что для 3 микроРНК (hsa-miR-17-5p, hsa-miR-141-5p, hsa-miR-556-3p) с пониженной экспрессией в эндометриоидной ткани, экспрессия регулируемых ими мРНК-транскриптов PTGIS, SORCS2 и AOC3 повышена. Для 4 микроРНК (hsa-miR-145-5p, hsa-miR-21-5p, hsa-miR-222-3p, hsa-miR-34c-5p) с повышенной экспрессией наблюдается снижение уровня экспрессии их генов-мишеней ESR1, TOP2A, ASRGL1, CDH1, SOX17. Уровень hsa-miR-200a-5p и hsa-miR-200b-5p, регулирующих ген SOX17 по данным литературы может как снижаться, так и повышаться, что требует дополнительного подтверждения их функциональной роли. Для hsa-miR-145-5p и hsa-miR-21-5p, регулирующих экспрессию FZD7 и TCF21 соответственно показано однонаправленное повышение как микроРНК, так и количества транскриптов. Приведенные данные могут служить дополнительным доказательством участия дифференциально экспрессированных микроРНК в патогенезе эндометриоза.
В связи с тем, что связывание микроРНК с мРНК не всегда приводит к изменению количества последней, для установления однозначного соответствия экспрессии микроРНК и мРНК генов-мишеней, необходимо проводить дополнительные лабораторные исследования с целью установления непосредственного взаимодействия микроРНК-мРНК, влияния микроРНК на содержание целевого белка в биообразце, а также влияния микроРНК на выполняемую белком функцию. Тем не менее, примененный аналитический подход может быть использован для выбора молекулярных объектов (микроРНК, мРНК, белков) для дальнейших исследований. Например при изучении экспрессии микроРНК при помощи микрочиповых технологий или высокопроизводительного секвенирования получается обширный список, состоящий из десятков или сотен дифференциально экспрессирующихся микроРНК. Валидация всех микроРНК из списка при помощи вышеуказанных подходов является трудновыполнимой и ресурсоемкой задачей. Поэтому перед исследователями встает проблема выбора кандидатов для дальнейшей валидации. В основе данного выбора лежит информация об уровне и направлении экспрессии кандидатных молекул, о ранее проведенных исследованиях по их валидации, а также информация о вовлечении кандидатов в физиологические процессы. Таким образом, сопоставление результатов, полученных в эксперименте, с результатами мета-анализа транскриптомных баз данных представляется оправданным подходом в решении данной проблемы. Кроме этого, информация об экспрессии микроРНК, как регулятора трансляции целевого белка, может быть дополнительным подтверждением его роли в патологическом процессе.
В научной литературе, посвященной патогенезу эндометриоза, показаны изменения экспрессии различных генов, кодирующих транскрипционные факторы, молекулы клеточной адгезии, ферменты тканевого ремоделинга, цитокины, рецепторы и ферменты метаболизма стероидных гормонов. Однако проведенный анализ публикаций показал, что для большинства описанных нами генов, отсутствует информация об экспрессии кодируемых ими белков в очагах эндометриоза. Основной массив данных о взаимосвязи между белковыми продуктами проанализированных генов и сигнальными путями, в которых они участвуют, получен при исследовании опухолевых или эмбриональных клеток. По данным проведенного анализа можно предложить несколько возможных кандидатов для дальнейших исследований. Представляется перспективным изучить выявленные компоненты Wnt-каскада, поскольку в данном исследовании обнаружено сразу четыре гена, ассоциированных с этим путем. При этом два из этих генов могут контролироваться микроРНК, изменение экспрессии которых обнаружено при эндометриозе. Изменение экспрессии генов эстрогенового рецептора ERa, ряда генов транскрипционных факторов и ферментов метаболизма эстрогенов, а также ассоциированных с ними микроРНК может позволить сформировать новые представления о гормональном контроле в эндометриоидной ткани. Таким образом, необходимы дальнейшие исследования с использованием «омиксных» и биоинформационных технологий, направленные на определение белковых продуктов генов, их участия в сигнальных путях и биологических процессах в клетках и тканях эктопического и эутопического эндометрия, для составления относительно полного интерактома сигнальных путей, участвующих в патогенезе эндометриоза, и выявления возможных терапевтических мишеней для лечения этого заболевания.



