Genotyping of embryos using fragmental STR analysis after whole genome amplification
Aim. To develop a method for determining the maternal or paternal origin of chromosomal aneuploidies in embryos by performing STR analysis for patients who undergo assisted reproductive technologies (ART) programs.Ekimov A.N., Alexandrova A.N., Alwexandrova N.V., Shubina E.S., Ritcher O.V., Goltsov A.Yu., Nazarenko T.A.
Materials and methods. The development of the method was carried out using biopsy material of the trophectoderm in embryos of 58 couples of patients (116 persons). Preimplantation genetic testing for aneuploidy (PGT-A) was performed using high-throughput sequencing. Determination of the origin of aneuploidy was performed by fragment analysis of STR markers.
Results. After After PGT-A, aneuploidy was detected in 67 embryos (41%); euploid embryos were 65 (40%); mosaic forms were found in 28 (17%) embryos. Primers have been developed for the analysis of chromosomes 1, 3, 4. Development of primers for analysis of 1, 3, 4, 5, 6, 7, 9, 11, 12, 13, 14, 15, 16, 19, 22 and X chromosomes was performed. The analysis of chromosome 16 (n = 7 embryos), chromosome 19 (n = 5 embryos) and chromosome 22 (n = 8 embryos) showed that this approach can be used to identify both maternal and paternal origin of aneuploidy.
Conclusion. A technological possibility of carrying out STR fragment analysis with purpose of genotyping of embryos using various products of whole genome amplification was shown. It makes possible to carry out PGT-A and PGT-M together with embryo genotyping without the need for repeated biopsies. The study parental contribution to the development of embryonic aneuploidy in future will allow to choose the most optimal tactics for managing patients with repeated unsuccessful attempts of ART and past miscarriages.
Keywords
A key step in each IVF cycle is the selection of the embryo with the highest implantation potential [1]. Nevertheless, transfer of an embryo with good morphology does not always lead to its implantation [2], since chromosomal aneuploidy (CA) is one of the factors of implantation failure and early reproductive losses [3]. In addition, aneuploidies that lead to a cessation of embryonic development and impaired cell division have been described [3]. At present, morphological methods for assessing the quality of an embryo do not allow detecting aneuploidy with a sufficiently high accuracy [4]. It was found that with an increasing mother age, the probability of detecting chromosomal abnormalities of embryos increases [5], however, even with fertilization of donor eggs obtained from young healthy women, CA of embryos are found in 18–61% of cases [6].
Most aneuploidies are of maternal origin, i.e. arise due to meiotic errors [7]. Previously, it was shown that in blastomeres, chromosomal trisomies of maternal origin were detected almost 10 times more often than trisomies of paternal origin. Preimplantation genetic testing for aneuploidy (PGT-A) using polar body analysis has shown that errors in meiosis II (MII) are more common than in meiosis I (MI). Errors of maternal meiosis are the main causes of the formation of CA in human embryos; 45% of cases CA among them are formed due to MII, and 34% of CA of embryos – due to MI [8].
In recent years, the proportion of patients of older reproductive age who seek treatment for infertility with the help of assisted reproductive technologies (ART) has significantly increased [9], while the detection of CA in them can be up to 90% of cases. Thereby lot of efforts are made to introduce new approaches to diagnose CA. Technologies that allow to evaluate all 24 chromosomes include comparative genomic hybridization [10], quantitative polymerase chain reaction (PCR) [11, 12], and high-throughput sequencing [13–15]. The disadvantage of the above methods is the inability to distinguish maternal or paternal origin of aneuploidies. The origin of aneuploidy can be determined using the detection and comparative analysis of single nucleotide polymorphisms (SNPs) in DNA samples of embryos and
their parents [7]. Genotyping both parents and the embryo at the same time allows us to distinguish the parental origins of each chromosome, as well as to identify the positions at which crossing over occurred. Moreover, using SNP-haplotyping, it is possible to measure the number of chromosomes. All this make it possible to obtain comprehensive information about the mechanisms of the appearance and origin of CA in embryos. However, this method is very expensive and requires special equipment.
Purpose of the study: to develop a method of determining the maternal or paternal origin of the CA of an embryo using STR analysis in patients with ART programs.
Materials and methods
To develop this method, we used material from a biopsy of embryo trophectoderm of 58 pairs of patients (116 people) who applied for an ART program with OGT-A for the following indications: the mother's age is over 35; repeated unsuccessful attempts at IVF in anamnesis (2 or more); habitual miscarriage; severe spermatogenesis disorders; patient's desire. The patients underwent an outpatient examination before the IVF program, which included mandatory research methods, special research methods, as well as studies for medical reasons. Informed consent was obtained from all patients to conduct this study. Blood samples were obtained from each pair, collected in tubes with EDTA for further DNA extraction.
A protocol with a gonadotropin-releasing hormone antagonist was used to stimulate ovarian function. Stimulation was carried out with preparations of recombinant FSH, a combined preparation of recombinant FSH and LH, or human menopausal gonadotropin from the 2nd or 3rd day of menstrual cycle. The dose of the drug depended on the patient's age and ovarian reserve (AMH level and the number of antral follicles according to ultrasound data). Mature oocytes obtained during transvaginal puncture were fertilized by ICSI. After fertilization the oocytes were transferred to Cook culture medium (Australia).
Morphological embryo assessment was carried out after 120–122 hours (on the 5th day) of cultivation. The morphological embryo characteristics were taken into account according to the Gardner classification (the degree of maturity of blastocysts, the quality of the intracellular mass, and the quality of the trophoectoderm (TFE). TFE biopsy was performed on the 5th day of embryo cultivation in vitro (120 hours after transvaginal puncture) using a Narishiga micromanipulator (Japan). A Fertilase laser gun (Germany) was used to dissect the zona pellucida. Cook micropipettes (Australia) were used for aspiration of TFE cells. An average of 5–10 TFE cells of each embryo were washed in sterile medium containing HEPES buffer (FertiPro) and placed in 3 μl drops in 0.2 ml Eppendorf PCR tubes.
The whole genome amplification was performed using WGA-PCR (Rubicon, USA) and MDA (Qiagen, USA). PGT-A was performed using high-throughput sequencing on ReproSeq kits (Thermo Fisher Scientific, USA). Subsequently, the whole genome amplification was used for fragment analysis. For this, an additional stage of amplification was carried out using the original fluorophore-labeled primers for STR analysis developed in this study. Fragmental analysis of parental DNA was performed without a preliminary stage of whole genome amplification. The resulting fluorophore-labeled amplicons were analyzed using a Genetic Analyzer 3130 capillary electrophoresis instrument (Applied Biosystems). The analysis and comparison of the obtained STR profiles were carried out using the GeneMapper ID v3.2.1 software (Applied Biosystems).
Results
In total, 167 embryos were used to develop this methodological approach. The average number of analyzed embryos per patient was 2.8.
After PGT-A, aneuploidies were detected in 67 embryos (41%), 65 (40%) were euploid, mosaic forms were found in 28 (17%) embryos. 3 embryos were polyploid. The whole genome amplification did not take place in 4 embryos (2%). Most often, aneuploidies in embryos were associated with 22 and 16 chromosomes (Fig. 1).
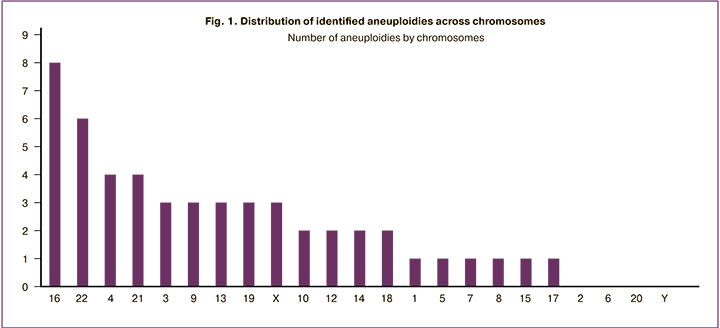
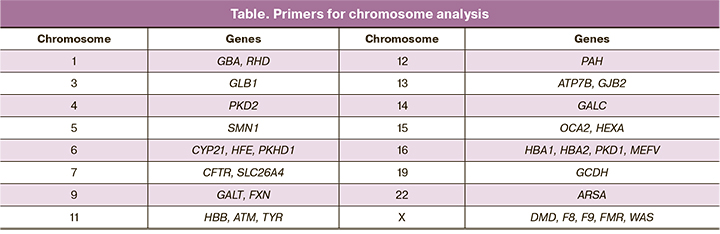
Within the framework of this study, primers were selected and synthesized for the analysis of chromosomes 1, 3, 4, 5, 6, 7, 9, 11, 12, 13, 14, 15, 16, 19, 22, and X (Table). The choice of these genes and regions was substantiated by the fact that the obtained STR markers were informative for carrying out the PGT-M of the corresponding diseases. For example, human chromosome 5 primers were designed to study the inheritance of diseases associated with disorders in the SMN1 gene (spinal muscular atrophy). Primers for the study of chromosome 16 are located in the region of the PKD1 gene and are used to study the inheritance of polycystic ovary syndrome (PCOS).
In order to determine the possibility of carrying this study out on various biological material, we worked out the fragment analysis on samples obtained by different methods: 1) whole genome amplification using WGA-PCR obtained using Rubicon kits (USA); 2) whole genome amplification obtained by WGA-PCR with barcodes for high throughput sequencing (ThermoFisher, USA); 3) whole genome amplification obtained using MDA (Qiagen, USA); 4) native DNA isolated from blood (father and mother).
It has been shown that for successful STR analysis and determination of the parental contribution to the development of embryonic aneuploidies, one can use both the product obtained using WGA-PCR (both with and without barcodes for high-throughput sequencing), as well as products obtained using MDA. At the same time, reliable results can be obtained only if the STR markers of the parents do not coincide, since the magnitude of the peak and the area under the curve can be influenced by the non-equilibrium whole genome amplification of different regions of different chromosomes. Also, when using whole genome amplification, allele loss is possible, which makes it difficult or even impossible to interpret the results correctly; however, this phenomenon was not identified in our work.
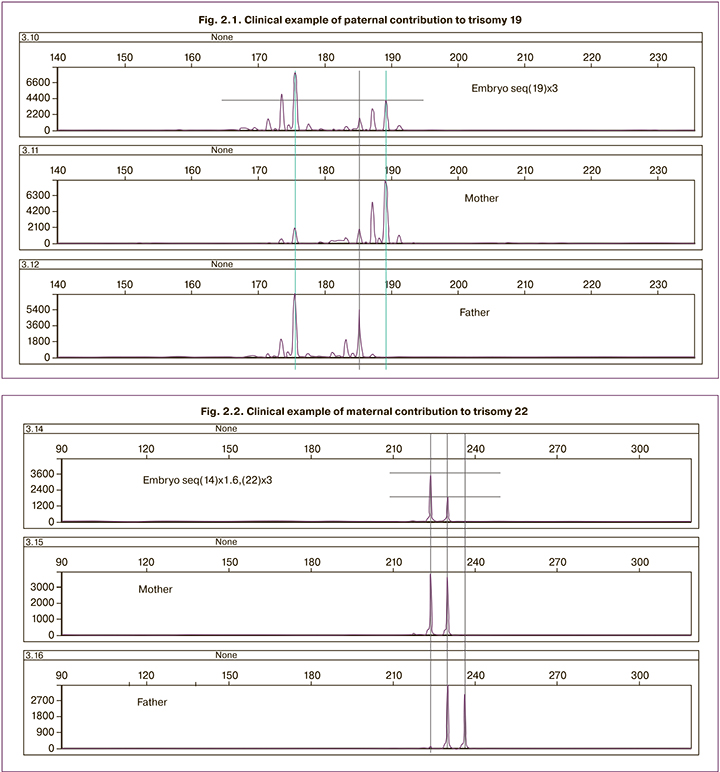
Fragmentary analysis of embryos for individual chromosomes (chromosome 16 (n=7 embryos), chromosome 19 (n=5 embryos) and chromosome 22 (n=8 embryos) showed that this approach can be used to identify aneuploidies, both of maternal and paternal origin (Fig. 2.1, 2.2). So in figure 2.1 it can be seen that the embryo received two 19 chromosomes from father and one from mother. In figure 2.2 it can be seen that the embryo received one chromosome 22 from the father and two from the mother.
Despite the fact that methods for identifying parental origin were developed for almost all aneuploidies, in a number of cases the interpretation of the results was ambiguous, which requires additional research in this area.
Discussion
During the analysis of 167 biopsy specimens of TFE of 58 patients who applied for an IVF program with OGT-A, aneuploid embryos were diagnosed in 41.0% of cases, which is consistent with previously obtained data [16, 17]. Of course, the maternal age plays an important role in the onset of aneuploidy; but aneuploidies in embryos are also detected in IVF programs with donor oocytes, when, as it is known, the donor's age does not exceed 35 years [6].
In our study, in 17% of cases, embryos with a mosaic genotype were identified; in case of transfer of these, the pregnancy rate was lower; this seemed to increase the risk of miscarriage [18]. The decision to transfer embryos with a mosaic genotype is currently a controversial issue without clear recommendations for a date. Therefore, we did not analyze such embryos in detail in this study.
The advantage of using STR fragment analysis is its relative simplicity and low cost. It requires a set of equipment, which is usually at the disposal of most genetic laboratories. This is a PCR amplifier and a capillary electrophoresis device. In addition, there is no need for expensive comparative genomic hybridization or high throughput sequencing instruments.
The analysis of embryos using fragment analysis of 16 (n=7 embryos), 19 (n=5 embryos) and 22 (n=8 embryos) chromosomes showed the fundamental possibility of determining the parental affiliation of chromosomes in aneuploid embryos.
When determining the tactics of treatment and examination of patients with repeated failures of IVF programs and miscarriage, including situation after carrying out OGT-A of embryos, the following scheme may be useful.
If, as a result of PGT-A with the determination of the parental origin of aneuploidies, there are no embryos recommended for transfer, it is advisable to repeat the program. In case of repeated absence of embryos for transfer according to the results of PGT-A with determination of the parental origin of aneuploidies, it is recommended to carry out an IVF program with donor cells. The choice of donor material (oocytes or sperm) is recommended based on the results of OGT: if the aneuploidies are of maternal origin – IVF program with donor oocytes, if paternal – IVF program using donor sperm.
If it is impossible to obtain embryos for the subsequent program with PGT-A with the determination of the parental origin of aneuploidies, it is recommended to focus on the program using donor cells.
Conclusion
The principal technological possibility of carrying out STR fragment analysis for the purpose of genotyping embryos using various products of whole genome amplification has been shown. This approach allows OGT-A and OGT to be performed for monogenic diseases in conjunction with embryo genotyping without the need for repeated biopsies, which can negatively affect their viability. The study of the parental contribution influence on the development of embryo aneuploidies in the future, possibly, will make it possible to choose the most optimal tactics for managing patients with repeated unsuccessful attempts of ART and a history of miscarriage.
References
- Faramarzi A., Khalili M.A., Ashourzadeh S. Oocyte morphology and embryo morphokinetics in an intra-cytoplasmic sperm injection programme. Is there a relationship? Zygote. 2017; 25(2):190-6. https://dx.doi.org/10.1017/S0967199417000041.
- El-Danasouri I., Sterzik K., Rinaldi L., Pacchiarotti A., DeSanto M., Selman H. Effect of transferring a morphologically impaired embryo with a good quality embryo on the pregnancy and implantation rates. Eur. Rev. Med. Pharmacol. Sci. 2016; 20(3): 394-8.
- Dahdouh E.M., Balayla J., García-Velasco J.A. Comprehensive chromosome screening improves embryo selection: a meta-analysis. Fertil. Steril. 2015; 104(6): 1503-12. https://dx.doi.org/10.1016/j.fertnstert.2015.08.038.
- Minasi M.G., Colasante A., Riccio T., Ruberti A., Casciani V., Scarselli F. et al. Correlation between aneuploidy, standard morphology evaluation and morphokinetic development in 1730 biopsied blastocysts: a consecutive case series study. Hum. Reprod. 2016; 31(10): 2245-54. https://dx.doi.org/10.1093/humrep/dew18.
- Бейк Е.П., Сыркашева А.Г., Долгушина Н.В. Эффективность программ вспомогательных репродуктивных технологий у пациенток позднего репродуктивного возраста. Гинекология. 2018; 20(1): 109-12. [Beik E.P., Syrkasheva A.G., Dolgushina N.V. Effectiveness of programs of auxiliary reproductive technologies in patients of late reproductive age. Gynecology. 2018; 20(1): 109-12. (in Russian)]. https://dx.doi.org/10.26442/2079-5696_20.1.109-112.
- Munné S., Alikani M., Ribustello L., Colls P., Martínez-Ortiz P.A., McCulloh D.N.; Referring Physician Group. Euploidy rates in donor egg cycles significantly differ between fertility centers. Hum. Reprod. 2017; 32(4): 743-9. https://doi.org/10.1093/humrep/dex031.
- Rabinowitz M., Ryan A., Gemelos G., Hill M., Baner J., Cinnioglu C. et al. Origins and rates of aneuploidy in human blastomeres. Fertil. Steril. 2012; 97(2): 395-401. https://dx.doi.org/10.1016/j. fertnstert.2011.11.034.
- Handyside A.H., Montag M. Multiple meiotic errors caused by predivision of chromatids in women of advanced maternal age undergoing in vitro fertilisation. Eur. J. Hum. Genet. 2012; 20(7): 742-7. https://dx.doi.org/10.1038/ejhg.2011.272.
- Назаренко Т.А. Эндокринные факторы женского и мужского бесплодия. Принципы гормонального лечения. М.: МИА; 2017. [Nazarenko T.A. Endocrine factors of female and male infertility. Principles of hormone treatment. Moscow, 2017. (in Russian)].
- Zhou Z., Ma Y.L., Li Q., Zhang Y., Huang Y.H., Tu Z.H. et al. Clinical application of oligo array-CGH for detecting balanced translocations in preimplantation genetic diagnosis. Int. J. Clin. Exp. Pathol. 2017; 10(7): 7821-35. eCollection 2017.
- Daser A., Thangavelu M., Pannell R., Forster A., Sparrow L., Chung G. et al. Interrogation of genomes by molecular copy-number counting (MCC). Nat Methods. 2006; 3(6): 447-53. https://dx.doi.org/10.1038/nmeth880.
- Treff N.R., Tao X., Ferry K.M., Su J., Taylor D., Scott R.T. Development and validation of an accurate quantitative real-time polymerase chain reaction-based assay for human blastocyst comprehensive chromosomal aneuploidy screening. Fertil. Steril. 2012; 97(4): 819-24. 10. https://dx.doi.org/1016/j.fertnstert.2012.01.115.
- Fiorentino F., Biricik A., Bono S., Spizzichino L., Cotroneo E., Cottone G. et al. Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil. Steril. 2014; 101(5): 1375-82. https://dx.doi.org/10.1016/j.fertnstert.2014.01.051.
- Aleksandrova N., Shubina E., Ekimov A., Kodyleva T., Mukosey I., Makarova N., Kulakova E., Levkov L., Trofimov D., Sukhikh G. Comparison of the results of preimplantation genetic screening obtained by a-CGH and NGS methods from the same. Gynecol. Endocrinol. 2016; 32(Suppl. 2): 1-4. https://dx.doi.org/10.1080/09513590.2016.1232892.
- Александрова Н.В., Шубина Е.С., Екимов А.Н., Кодылева Т.А., Мукосей И.С., Макарова Н.П., Кулакова Е.В., Левков Л.А., Барков И.Ю., Трофимов Д.Ю., Сухих Г.Т. Молекулярная биология. 2017; 51(2): 308-13. [Aleksandrova N.V.,Shubina E.S., Ekimov A.N., Kodyleva T.A., Mukosey I.S., Makarova N.P.,Kulakova E.V., Levkov L.A., Barkov I.Y., Trofimov D.Y., Sukhikh G.T. Comparative results of preimplantation genetic screening by array comparative genomic hybridization and new-generation sequencing. Mol. Biol. 2017; 51(Suppl. 2): 308-13. (in Russian)]. https://dx.doi.org/10.7868/S0026898417010025.
- Demko Z.P., Simon A.L., McCoy R.C., Petrov D.A., Rabinowitz M. Effects of maternal age on euploidy rates in a large cohort of embryos analyzed with 24-chromosome single-nucleotide polymorphism–based preimplantation genetic screening Fertil. Steril. 2016; 105(5): 1307-13. https://dx.doi.org/10.1016/j.fertnstert.2016.01.025.
- McCoy R.C., Demko Z.P., Ryan A., Banjevic M., Hill M., Sigurjonsson S. et al. Evidence of selection against complex mitotic-origin aneuploidy during preimplantation development. PLOS Genet. 2015; 11(10): e1005601. https://dx.doi.org/10.1371/journal.pgen.1005601.
- Fragouli E., Wells D. Aneuploidy in the human blastocyst. Cytogenet. Genome Res. 2011; 133(2-4): 149-59. https://dx.doi.org/10.1159/000323500.
Received 15.12.2020
Accepted 24.12.2020
About the Authors
Alexey N. Ekimov, clinical geneticist of Molecular Genetics Laboratory, V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia. Tel.: +7(495)531-44-44. Е-mail: a_ekimov@oparina4.ru. 4, Oparina str., Moscow, 117997, Russian Federation.Natalia V. Aleksandrova, M.D., leading researcher, V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia. Tel.: +7(495)531-44-44. Е-mail: alexnat1@yandex.ru. 4, Oparina str., Moscow, 117997, Russian Federation.
Ekaterina S. Shubina, Head of Genomic Data Analysis Laboratory, V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia. Tel.: +7(495)531-44-44. Е-mail: e_shubina@oparina4.ru. 4, Oparina str., Moscow, 117997, Russian Federation.
Andrey Yu. Goltsov, researcher at Molecular Genetics Laboratory, V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia. Tel.: +7(495)531-44-44. Е-mail: andrey.goltsov@gmail.com.4, Oparina str., Moscow, 117997, Russian Federation.
Olga V. Ritcher, clinical laboratory diagnostician of Molecular Genetics Laboratory, V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia. Tel.: +7(495)531-44-44. Е-mail: o_ritcher@oparina.ru.4, Oparina str., Moscow, 117997, Russian Federation.
Tatiana A. Nazarenko, Dr. Med. Sci., professor, Head of Institute of Reproductive Medicine, V.I. Kulakov National Medical Research Center for Obstetrics, Gynecology and Perinatology, Ministry of Health of Russia. Tel.: +7(495)531-44-44. Е-mail: t_nazarenko@oparina4.ru. 4, Oparina str., Moscow, 117997, Russian Federation.
For citation: Ekimov A.N., Alexandrova A.N., Alwexandrova N.V., Shubina E.S., Ritcher O.V.,Goltsov A.Yu., Nazarenko T.A. Genotyping of embryos using fragmental STR analysis after whole genome amplification.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2021; 1: 126-132 (in Russian)
https://dx.doi.org/10.18565/aig.2021.1.126-132



