Sialidase activity of vaginal bacteria in reproductive-age women
Bacterial sialidase activity catalyzes the degradation of protective mucosal barriers.Shipitsyna E.V., Korkina S.A., Krysanova A.A., Kolousova K.A., Shalepo K.V., Budilovskaya O.V., Khusnutdinova T.A., Savicheva A.M., Kogan I.Yu.
Objective: To investigate sialidase activity of vaginal bacteria and identify sialidase genotypes and phenotypes of different species of Gardnerella spp.
Materials and methods: A semiquantitative sialidase assay was developed and used to test 89 strains of vaginal bacteria, including 19 strains of Gardnerella spp. The Gardnerella spp. species were identified by DNA sequencing. Gardnerella spp. sialidase genes were detected by PCR.
Results: Sialidase activity was detected in 8 strains belonging to three genera, including Anaerococcus (n=1), Bifidobacterium (n=1), and Gardnerella (n=6). G. vaginalis represented the majority of the Gardnerella strains (n=16, 84%); the proportion of G. swidsinskii was (n=3, 16%). Nine strains of G. vaginalis had only the nanH1 gene and all of these strains did not exhibit sialidase activity. The remaining seven strains of G. vaginalis simultaneously had the nanH1 and nanH3 genes, and sialidase activity was registered in 6 strains. No sialidase genes were detected in any G. swidsinskii strain.
Conclusion: Most cultured species of vaginal bacteria, including lactobacilli, do not produce sialidase activity. The predominant species of the genus Gardnerella is G. vaginalis (84%); the proportion of G. swidsinskii is 16%. Bifidobacterium bifidum, Anaerococcus tetradius strains and most strains of G. vaginalis, whose genome contains both the nanH1 and nanH3 genes, can degrade sialoglicans. G. swidsinskii strains, as well as G. vaginalis strains with only the nanH1 gene, do not exhibit sialidase activity.
Keywords
Cervical mucus isolates the uterus from the vaginal compartment and thereby protects the upper genital tract from microbial invasion. In addition to its protective function, cervical mucus lubricates the lower genital tract and, due to its high carbohydrate content, can serve as energy and nutrient source for resident bacterial populations of the vagina and cervical canal.
The main components of mucous gels, including cervical mucus, are mucins, large, highly glycosylated glycoproteins. The carbohydrate side chains are thought to protect the underlying protein backbone from proteolysis. Mucins are produced by goblet cells of mucous membranes and submucosal glands. In addition to mucins, mucin gels contain inorganic salts, antimicrobial components such as lysozyme, lactoferrin, and immunoglobulins.
Mucinases are enzymes produced by some microorganisms and viruses and are capable of degrading mucin. The main group of mucinases are glycosidases, that include the best-studied enzyme sialidase, or neuraminidase. Sialidases remove terminal sialic acid residues from the glycan chain, thereby modulating various cellular processes, both physiological and pathological. Thus, some bacteria resident normally produce small amounts of mucinase, thus contributing to the normal process of mucus renewal [1]. At the same time, partial or complete degradation of mucin molecules by mucinases is often a critical step in the destruction of mucosal protective barriers.
The role of mucinase activity of microorganisms and viruses in the pathogenesis of various infections has been studied for several decades, but research has mainly focused on infections of the gastrointestinal and respiratory tracts. Information on the role of vaginal bacterial mucinases in the pathogenesis of reproductive tract infections is fragmentary and insufficient. However, the production of mucinases by vaginal bacteria may be one of the main microbiological factors of ascending infection [2, 3], which can lead to severe diseases of the female reproductive tract including pelvic inflammatory diseases, infertility, and ectopic pregnancy. During pregnancy, the ascending infection can cause miscarriage and preterm birth.
The mucosal surfaces of the female urogenital tract are particularly rich in sialidases. Sialidases of some bacteria catalyze removal of these residues thereby promoting colonization [4]. Bacterial vaginosis (BV) associated bacteria such as Gardnerella vaginalis play a key role in BV pathogenesis, are considered to be the main producers of mucolytic enzymes in the vagina [5]. Gardnerella are thought to use sialidase to degrade and deplete components of vaginal mucus [5]. The genus Gardnerella until recently included a single species, G. vaginalis. However, given the significant biotypic diversity of G. vaginalis, including the role of sialidase production, numerous attempts have been made to genotype these bacteria, including by high-throughput sequencing. In 2019, full-genome sequence analysis was performed for 81 Gardnerella genomes, resulting in the addition of newly described species Gardnerella leopoldii, Gardnerella piotii and Gardnerella swidsinskii [6]. Because of the high 16SrRNA sequence similarity (98.5%), the Gardnerella species are currently identified by sequencing the 60 kDa chaperonin gene [7].
Until recently, it was thought that the sialidase activity of Gardnerella spp. was associated with the sialidase A gene, later named nanH1. Recently, two more sialidases, NanH2 and NanH3, were discovered in Gardnerella spp. This study showed that NanH2 and NanH3 were more efficient than NanH1 in the cleavage of sialic acids on mucosal substrates. Further, the presence of the nanH1 sialidase gene has not always been associated with detectable sialidase activity [4].
This study aimed to investigate sialidase activity of vaginal bacteria and identify sialidase genotypes and phenotypes of different species of Gardnerella spp.
Materials and methods
Culture study of vaginal discharge samples
The study analyzed samples of vaginal discharge obtained from women of reproductive age at the Ott Research Institute of Obstetrics, Gynecology, and Reproductology in 2021–2022. Clinical material was inoculated on Schedler agar designed to culture facultative anaerobes and anaerobes and incubated for 48 h at 37°C and 5% CO2 concentration in an anaerobic chamber using gas packets (AnaeroGen, Oxoid, UK).
To purify the colonies of all morphotypes grown on Schedler's agar, they were dissected three times by depleting smear and identified by mass spectrometry (MALDI-TOF MS, Bruker Daltonics, Germany). This method allows identification of microorganisms based on comparison of the total protein mass spectrum with a database of reference strains. Only bacterial strains for which a high reliability of identification was noted (the value of the similarity index of the spectra of the tested and reference strains was greater than 2.000) were selected for the study. The identified strains were suspended in 1.5 ml of tryptone-soy broth, frozen, and stored at -80°C. A total of 89 bacterial strains from 45 women were obtained (1–3 strains from each woman).
Semi-quantitative analysis of the sialidase activity of vaginal bacteria
A modification of the Robinson L.S. et al. (2019) technique was developed for semi-quantitative analysis of sialidase activity of vaginal bacteria [4]. Before analysis, bacterial cultures were thawed and, if necessary, concentrated by centrifugation to an optical density of 1.0 at 620 nm. A 100 mM sodium acetate solution containing 300 μM 2-(4-methylumbelliferyl)-α-D-D-N-acetylneuraminic acid (MUAN) (Sigma-Aldrich, USA) was used as a fluorescent substrate. 80 μl of 100 mM sodium acetate solution containing 300 μM MUAN was added to the microplate wells for fluorescence. Then, 20 μl of a suspension of the bacterial strain under study and 20 μl of each dilution of the positive control were added to the wells, Clostridium perfringens sialidase (Sigma-Aldrich, USA) at concentrations of 0.001 iu/μL (K1, 0.02 iu/reaction), 0.0005 iu/μL (K2, 0.01 iu/reaction), 0.00025 iu/μL (K3, 0.005 iu/reaction), 0.000125 (K4, 0.0025 iu/reaction) were used as a positive control. A 100 mM sodium acetate solution was used as a negative control. The microplate was incubated at 37°C for 1 h, after which reaction results were visualized in a transilluminator (VilberLourmat, France) using VisionCapt software (VilberLourmat, France). The results of the analysis were recorded by comparing the fluorescence intensities of the strains under study and the controls. Positive controls conditionally corresponded to strongly pronounced (++++), pronounced (+++), moderate (++), and weakly pronounced (+) sialidase activity (K1-4, respectively).
Extraction of genomic DNA from cultures of Gardnerella spp.
Commercial DNA extraction kits AmpliPrime DNA-sorb-AM (InterLabService, Russia) were used to isolate DNA from Gardnerella spp. cultures for subsequent species and sialidase genotype identification. Gardnerella spp. cultures were thawed, after which DNA was extracted according to the manufacturer's recommended protocol.
Identification of Gardnerella spp. species and determination of their sialidase genotypes
Species identification of Gardnerella spp. was performed using Sanger sequencing of the chaperonin cpn60 gene [8] in a Beckman Coulter genetic analysis system, USA. First, the cpn60 gene was amplified by polymerase chain reaction (PCR) using Tersus plus PCR kit (Evrogen, Russia) and primers H729 (5'-CGCCAGGGTTTTCCCAGTCACGACGAIIIIGCIGGIGAYGGIACIACIAC-3') and H730 (5'-AGCGGATAACAATTTCACACAGGAYKIYKITCICCRAAICCIGGIGCYTT-3') (Eurogen, Russia) in the following thermal cycling mode: 94°C – 5 min. ; 94°C – 30 s, 50°C – 30 s, 72°C – 45 s. (40 cycles); 72°C – 10 min. [9]. The amplicons obtained were analyzed by electrophoresis in 1% agarose gel for 40 min. DNA was isolated from the PCR mixture using AMPure reagent kits (Beckman Coulter) according to the manufacturer's protocol. Sequencing PCR was performed in a PalmCycler thermal cycler (Corbett Research, Australia) using each sequencing primer (sequences of sequencing primers H729-M13 and H730-M13 are underlined) using the DTCS QuickStart kit (Beckman Coulter) according to the manufacturer's protocol with the following cycling parameters: 96°C – 20 s, 50°C – 20 s, 60°C – 4 min (30 cycles). PCR products were purified using CleanSeq reagent kits (Beckman Coulter) according to the manufacturer's protocol. Capillary electrophoresis was performed in a Beckman Coulter genetic analyzer. A consensus sequence was obtained using Investigator software (Beckman Coulter), which was then analyzed in the cpnDB database (https://www.cpndb.ca/).
Characterization of sialidase genotypes of Gardnerella spp.
Gardnerella spp. strains were analyzed for nanH1, nanH2, and nanH3 genes by PCR using primers and conditions developed by Robinson L.S. et al. (2019) [4] (Table 1). Tersus plus PCR kit (Eurogen) and Tertsik thermal cycler (DNA-Technology, Russia) were used for PCR amplification.
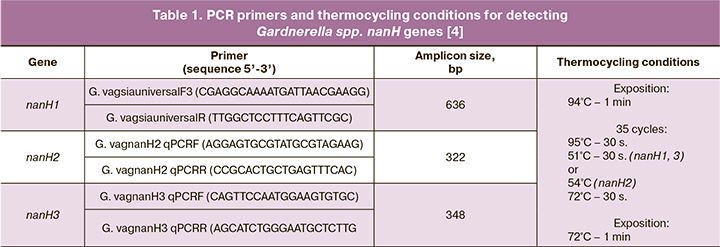
The obtained amplicons were separated on a 2% agarose gel. The results were visualized using an Infinity gel documentation system (VilberLourmat, France).
Results
The collection of vaginal bacteria consisted of 89 strains and included representatives of 42 species from 18 genera. All strains were analyzed by a semiquantitative method for the expression of sialidase activity (Figure). All strains of Gardnerella spp. and sialidase-positive cultures of other bacterial species were retested and all results were fully reproduced.
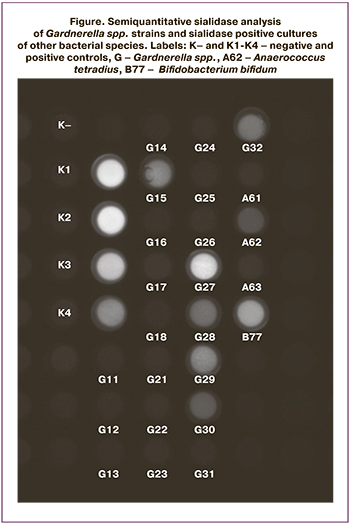
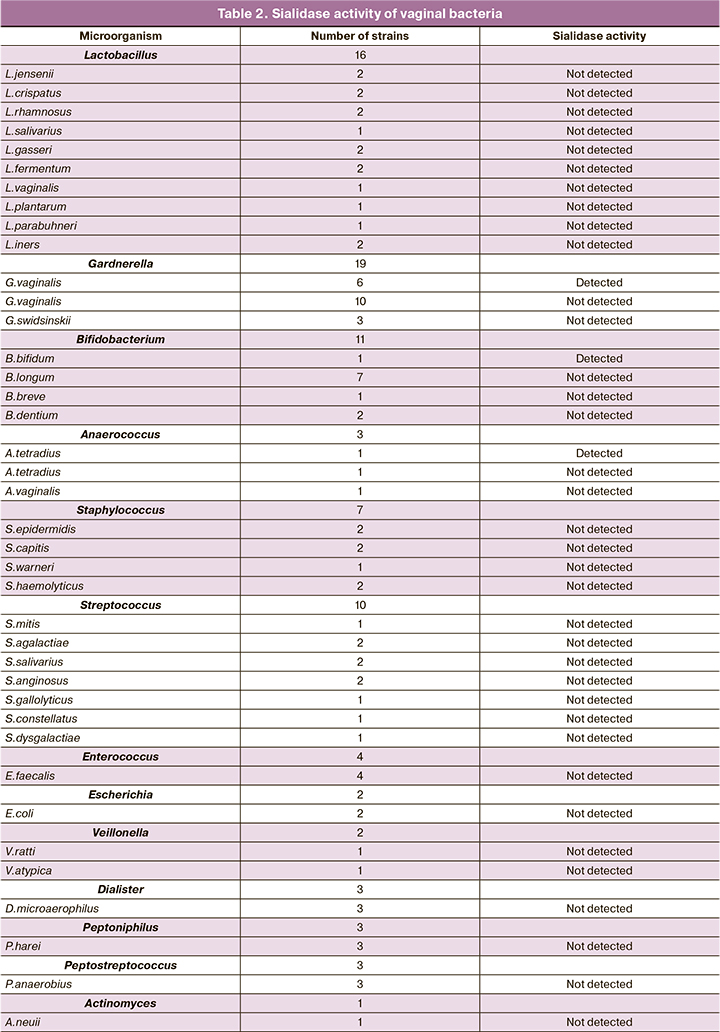
Most of the collection strains (81 out of 89 strains) did not produce sialidase activity (Table 2). Sialidase activity was detected in 8 strains belonging to three genera, including Anaerococcus (n=1), and Bifidobacterium (n=1), Gardnerella (n=6). Among 3 isolated strains of Anaerococcus spp. 2 strains were identified as A. tetradius (formerly Peptostreptococcus tetradius) and 1 as A. vaginalis (formerly P. vaginalis). Weak sialidase activity was noted in one of the two, A. tetradius; A. vaginalis was sialidase-negative. The genus Bifidobacterium was represented by the species B. bifidum, B. breve, B. dentium, and B. longum. Moderate sialidase activity was detected only in the B. bifidum strain. Of the 19 strains of Gardnerella spp. 6 strains had sialidase activity: 1 strong sialidase activity, 2 moderate sialidase activity, and 3 weak sialidase activity.
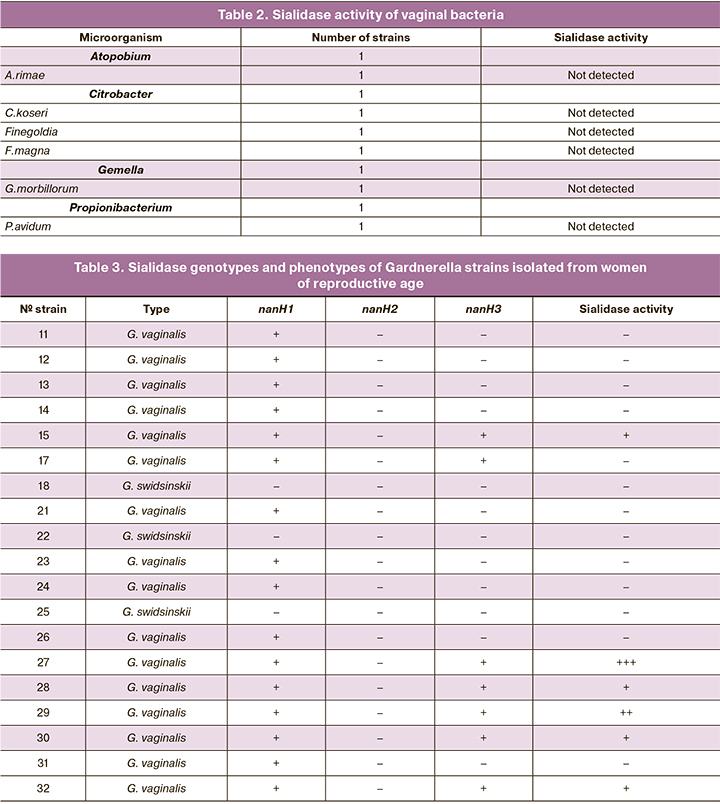
Sequencing of the cpn60 gene allows an identification of the species of the Gardnerella strains. Three strains (16%; 95% CI 4–40%) were identified as G. swidsinskii, the remaining 16 (84%; 95% CI 60–96%) as G. vaginalis. The 19 strains of Gardnerella spp. strains were analyzed for the presence of sialidase genes nanH1, nanH2 and nanH3 sialidase genes, and the results were compared with those of the sialidase activity test (Table 3). None of the three strains of G. swidsinskii possessed sialidase genes, and all 3 strains were sialidase negative. In 9 strains of G. vaginalis, only the nanH1 gene was detected; all of these strains also did not show sialidase activity. The remaining 7 strains of G. vaginalis, the nanH1 and nanH3 genes were detected simultaneously and sialidase activity was registered in 6 strains and not in 1 strain.
Discussion
The ability of some microorganisms and viruses to ferment glycans (primarily sialylglycans) as a factor in the infection process has been studied mainly in infections of the respiratory and gastrointestinal tracts. In respiratory infections, the vast majority of studies have focused on sialidase from influenza viruses. In intestinal infections, this enzymatic ability has been described in representatives of the Bacteroidetes, Actinobacteria, and Firmicutes types that dominate the intestinal microbiota. While some intestinal bacteria use mucin glycans only as adhesion sites, other bacteria, in addition, utilize them as a nutrient substrate [10].
The ability of most strains of Bastegoides bacteria to ferment glycans has been fairly well characterized [11, 12]. Bacteroides includes bacterial species widely represented in the human microbiota (including the vaginal microbiota) of the genera Bacteroides, Porphyromonas and Prevotella. Genomic and functional studies show that bacteroides prefer complex carbohydrates over simple carbohydrates, including mucin glycans [13].
At the same time, it is shown, also on the example of the intestinal microbiota, that most Firmicutes-type bacteria prefer the assimilation of simpler carbohydrates rather than complex carbohydrates [14]. When applied to the vaginal microbiota, Firmicutes include Lactobacillus spp., the dominant bacteria of the normal vaginal microbiota. Some species of Lactobacillus spp. are endowed with mucin-binding proteins [13], but they apparently lack any mechanism for its utilization. In this study, we have shown for the first time, in a number of strains of different species of vaginal lactobacilli, that these bacteria do not produce sialidase activity. Firmicutes type also includes streptococci, staphylococci, enterococci, and bacteria of the order Clostridiales. We also detected no sialidase activity in Streptococcus, Staphylococcus, and Enterococcus strains. It should be noted that several pathogenic and conditionally pathogenic Firmicutes, such as sialidase-producing Streptococcus pneumoniae, use the mechanism of glycan fermentation to adhere to the epithelium of the respiratory or intestinal tract [15]. Clostridiales in our collection were represented by bacteria of the genera Veillonella, Dialister, Peptoniphilus, Peptostreptococcus, and Anaerococcus. Of the three strains of Anaerococcus spp. (2 strains of A. tetradius and 1 strain of A. vaginalis), one of the two strains of A. tetradius showed sialidase activity; the strain of A. vaginalis was sialidase-negative. A. tetradius bacteria were classified as Peptostreptococcus tetradius until recently, and in one of the early works had a negative sialidase phenotype [12]. It is likely that not all strains of this species have sialidase genes or sialidase genes have inducible rather than constitutive expression.
The ability to utilize glycans is also inherent in several representatives of the Actinobacteria type, which include bacteria of the Bifidobacteriaceae family. Bifidobacteriaceae include bacteria widely represented in the human microbiota, such as Bifidobacterium spp. – symbiont gut bacteria that dominate the intestinal microbiota of the infant, and Gardnerella spp., detected in the vaginal microbiota of both healthy women (in small numbers) and women with BV (as the dominant species). In previous studies, the highest sialidase activity was detected in B. bifidum [16]; it was also shown that this species contains a large set of mucinases, including sialidase [17]. In our collection of vaginal bacteria, bifidobacteria were represented by B. bifidum, B. breve, B. dentium, and B. longum species, and sialidase activity was detected only in the B. bifidum strain.
In this study, 3 of the 19 Gardnerella strains were identified as G. swidsinskii and the remaining 16 were identified as G. vaginalis. The 3 G. swidsinskii strains were negative for sialidase genes and therefore for sialidase activity. In 9 strains of G. vaginalis, only the nanH1 gene was detected, and all of these strains were sialidase negative. The remaining 7 strains of G. vaginalis, the nanH1 and nanH3 genes were detected simultaneously and sialidase activity was recorded in all but one of these strains. The data we obtained are consistent with the results of recent studies [4, 9], which showed that the presence of the nanH3 gene in addition to the nanH1 gene was necessary for the expression of sialidase activity. Furthermore, in these studies, the same pattern was found for the nanH2 gene, and the frequency of this gene was very low. In our study, the nanH2 gene was not detected in any strain, which is also consistent with the results of previous studies [4, 9].
Studies investigating the role of sialidase-producing bacteria in infectious processes in the female urogenital tract are mainly limited to the pathogenesis and diagnosis of BV [12, 18, 19]. The first work on the subject [12] showed that women with BV had significantly higher concentrations of sialidase activity products than women without BV. The authors also examined the sialidase activity of several vaginal bacterial strains and found that all Prevotella bivia isolates, 40% of P. disiens isolates and 20% of G. vaginalis produced sialidase, while none of the isolates of Mobiluncus curtisii, M. mulieris, Peptostreptococcus spp. and Mycoplasma hominis showed sialidase production [12]. Subsequently, increased sialidase production in BV was shown in several other studies [18–20]. It is interesting to note that increased sialidase activity in vaginal discharge has been detected not only in women with BV, but also in women with aerobic vaginitis (AV), which is characterized by the dominance of Escherichia coli, Streptococcus spp., Staphylococcus spp. and Enterococcus spp. [21]. However, in our study, none of the strains of Escherichia coli, Streptococcus, Staphylococcus spp. and Enterococcus spp. had sialidase activity. An indirect confirmation of our results can be found in a study by Santos-Greatti et al. (2016) [22], which demonstrated significantly higher levels of sialidase activity in vaginal samples in women with BV microbiota than in women with intermediate Nugent microbiota, which, according to some reports, represents the AV microbiota [23]. Given the adverse effect of AV on female reproductive health [24], this issue deserves further investigation.
The clinical significance of the ability of bacteria to degrade mucins is far from limited to the significance of BV and AV. To date, sporadic studies have been published showing an association between mucinase (mainly sialidase) activity of vaginal bacteria and adverse pregnancy outcomes [25, 26]. The mechanisms by which mucolytic enzymes contribute to ascending infection are not fully understood. It is believed that degradation of protective mucus contributes to bacterial attachment to epithelial cells, altering the immune response and increasing the risk of spreading infection to the uterus. Once in the uterus, the bacteria continue to modulate the immune response, inducing a decrease in collagen synthesis in the amnion and chorion, as well as in the cervical canal tissue, and increasing the risk of premature rupture of membranes and preterm birth [26]. It is believed that the action of mucolytic microbial enzymes may also be a key step in the development of ascending infection in non-pregnant women, leading to infections of the upper reproductive tract (URT), perihepatitis, and postoperative complications. However, to date, the relationship between vaginal bacterial mucinase activity and URT infection has been largely unexplored.
Conclusion
Most cultured species of vaginal bacteria do not produce sialidase activity. The predominant species of Gardnerella spp. in the examined population is G. vaginalis (84%); the proportion of G. swidsinskii is 16%. Bifidobacterium bifidum, Anaerococcus tetradius strains and most strains of G. vaginalis, whose genome contains both the nanH1 and nanH3 genes, can degrade sialoglicans. G. swidsinskii strains and G. vaginalis strains with only the nanH1 gene do not exhibit sialidase activity.
References
- Glanz V.Y., Myasoedova V.A., Grechko A.V., Orekhov A.N. Sialidase activity in human pathologies. Eur. J. Pharmacol. 2019; 842: 345-50.https://dx.doi.org/10.1016/J.EJPHAR.2018.11.014.
- Vagios S., Mitchell C.M. Mutual preservation: a review of interactions between cervicovaginal mucus and microbiota. Front. Cell. Infect. Microbiol. 2021; 11: 676114. https://dx.doi.org/10.3389/fcimb.2021.676114.
- Lacroix G., Gouyer V., Gottrand F., Desseyn J.L. The cervicovaginal mucus barrier. Int. J. Mol. Sci. 2020; 21(21): 8266. https://dx.doi.org/10.3390/ijms21218266.
- Robinson L.S., Schwebke J., Lewis W.G., Lewis A.L. Identification and characterization of NanH2 and NanH3, enzymes responsible for sialidase activity in the vaginal bacterium Gardnerella vaginalis. J. Biol. Chem. 2019; 294(14): 5230-45. https://dx.doi.org/10.1074/jbc.RA118.006221.
- Agarwal K., Lewis A.L. Vaginal sialoglycan foraging by Gardnerella vaginalis: mucus barriers as a meal for unwelcome guests? Glycobiology. 2021; 31(6):667-80. https://dx.doi.org/10.1093/glycob/cwab024.
- Vaneechoutte M., Guschin A., Van Simaey L., Gansemans Y., Van Nieuwerburgh F., Cools P. Emended description of Gardnerella vaginalis and description of Gardnerella leopoldii sp. nov., Gardnerella piotii sp. nov. and Gardnerella swidsinskii sp. nov., with delineation of 13 genomic species within the genus Gardnerella. Int. J. Syst. Evol. Microbiol. 2019; 69(3): 679-87.https://dx.doi.org/10.1099/ijsem.0.003200.
- Hill J.E., Albert A.Y.K. Resolution and co-occurrence patterns of Gardnerella leopoldii, Gardnerella swidsinskii, Gardnerella piotii and Gardnerella vaginalis within the vaginal microbiome. Infect. Immun. 2019; 87(12): e00532-19. https://dx.doi.org/10.1128/IAI.00532-19.
- Vancuren S.J.., Hill J.E. Update on cpnDB: a reference database of chaperonin sequences. Database (Oxford). 2019; 2019: baz033. https://dx.doi.org/10.1093/DATABASE/BAZ033.
- Maier T., Pleckaityte M. Discrimination of Gardnerella species by combining MALDI-TOF protein profile, chaperonin cpn60 sequences, and phenotypic characteristics. Pathogens. 2021; 10(3): 277. https://dx.doi.org/10.3390/pathogens10030277.
- Tailford L.E., Crost E.H., Kavanaugh D., Juge N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015; 6: 81. https://dx.doi.org/10.3389/FGENE.2015.00081.
- Moncla B.J., Braham P., Hillier S.L. Sialidase (neuraminidase) activity among gram-negative anaerobic and capnophilic bacteria. J. Clin. Microbiol. 1990; 28(3): 422-5. https://dx.doi.org/10.1128/JCM.28.3.422-425.1990.
- Briselden A.M., Moncla B.J., Stevens C.E., Hillier S.L. Sialidases (neuraminidases) in bacterial vaginosis and bacterial vaginosis-associated microflora. J. Clin. Microbiol. 1992; 30(3): 663-6. https://dx.doi.org/10.1128/jcm.30.3.663-666.1992.
- van Tassell M.L., Miller M.J. Lactobacillus adhesion to mucus. Nutrients. 2011; 3: 613-36. https://dx.doi.org/10.3390/NU3050613.
- Ravcheev D.A., Thiele I. Comparative genomic analysis of the human gut microbiome reveals a broad distribution of metabolic pathways for the degradation of host-synthetized mucin glycans and utilization of mucin-derived monosaccharides. Front. Genet. 2017; 8: 11. https://dx.doi.org/10.3389/FGENE.2017.00111.
- Parker D., Soong G., Planet P., Brower J., Ratner A.J., Prince A. The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect. Immun. 2009; 77(9): 3722-30. https://dx.doi.org/10.1128/IAI.00228-09.
- Thum C., Roy N.C., McNabb W.C., Otter D.E., Cookson A.L. In vitro fermentation of caprine milk oligosaccharides by bifidobacteria isolated from breast-fed infants. Gut Microbes. 2015; 6(6): 352-63. https://dx.doi.org/10.1080/19490976.2015.1105425.
- Milani C., Turroni F., Duranti S., Lugli G.A., Mancabelli L., Ferrario C. et al. Genomics of the genus bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl. Environ. Microbiol. 2015; 82(4): 980-91. https://dx.doi.org/10.1128/AEM.03500-15.
- Smayevsky J., Canigia L.F., Lanza A., Bianchini H. Vaginal microflora associated with bacterial vaginosis in nonpregnant women: reliability of sialidase detection. Infect. Dis. Obstet. Gynecol. 2001; 9(1): 17-22. https://dx.doi.org/10.1155/S1064744901000047.
- Moncla B.J., Chappell C.A., Mahal L.K., Debo B.M., Meyn L.A., Hillier S.L. Mucinase and sialidase activity of the vaginal microflora: implications for the pathogenesis of preterm labour. Int. J. STD AIDS. 1999; 10(7): 442-7.https://dx.doi.org/10.1258/0956462991914438.
- Cauci S., Driussi S., Monte R., Lanzafame P., Pitzus E., Quadrifoglio F. Immunoglobulin A response against Gardnerella vaginalis hemolysin and sialidase activity in bacterial vaginosis. Am. J. Obstet. Gynecol. 1998; 178(3): 511-5. https://dx.doi.org/10.1016/S0002-9378(98)70430-2.
- Marconi C., Donders G., Bellen G., Brown D., Parada C., Silva M. Sialidase activity in aerobic vaginitis is equal to levels during bacterial vaginosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2013; 167(2): 205-9.https://dx.doi.org/10.1016/j.ejogrb.2012.12.003.
- Santos-Greatti M.M. de V., da Silva M.G., Ferreira C.S.T., Marconi C. Cervicovaginal cytokines, sialidase activity and bacterial load in reproductive-aged women with intermediate vaginal flora. J. Reprod. Immunol. 2016; 118: 36-41. https://dx.doi.org/10.1016/j.jri.2016.08.005.
- Donders G.G.G., Bellen G., Grinceviciene S., Ruban K., Vieira-Baptista P. Aerobic vaginitis: no longer a stranger. Res. Microbiol. 2017; 168(4): 845-58.https://dx.doi.org/10.1016/j.resmic.2017.04.004.
- Donders G.G.G., Bellen G., Rezeberga D. Aerobic vaginitis in pregnancy. BJOG. 2011; 118(10): 1163-70. https://dx.doi.org/10.1111/j.1471-0528.2011.03020.x.
- Howe L., Wiggins R., Soothill P.W., Millar M.R., Horne P..J., Corfield A.P. Mucinase and sialidase activity of the vaginal microflora: Implications for the pathogenesis of preterm labour. Int. J. STD AIDS. 1999; 10(7): 442-7.https://dx.doi.org/10.1258/0956462991914438.
- Cauci S., Culhane J.F. High sialidase levels increase preterm birth risk among women who are bacterial vaginosis positive in early gestation. Am. J. Obstet. Gynecol. 2011; 204(2): 142.e1-142.e9. https://dx.doi.org/10.1016/j.ajog.2010.08.061.
Received 14.07.2022
Accepted 25.08.2022
About the Authors
Elena V. Shipitsyna, Dr. Bio. Sci., Leading Researcher, International Department, D.O. Ott Reasearch Institute of Obstetrics, Gynecology and Reproductology,+7(812)323-75-44, shipitsyna@inbox.ru, 199034, Russia, St. Petersburg, Mendeleyevskaya Line, 3.
Sofia A. Korkina, student at the Department of Microbiology, Saint-Petersburg State University (199178, Russia, St. Petersburg, 16-th Line, 29),
performing final qualification work at the Department of Medical Microbiology, D.O. Ott Reasearch Institute of Obstetrics, Gynecology and Reproductology
(199034, Russia, St. Petersburg, Mendeleyevskaya Line, 3), ksa53@outlook.com
Anna A. Krysanova, Researcher, Department of Medical Microbiology, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology,
199034, St. Petersburg, Mendeleevskaya line, 3; Assistant, Department of Clinical Laboratory Diagnostics, St. Petersburg State Pediatric Medical University,
Ministry of Health of Russia, 194100, Russia, St. Petersburg, Litovskaya str., 2, 2474151@mail.ru
Kseniya A. Kolousova, student at the Department of Microbiology, Saint-Petersburg State University (199178, Russia, St. Petersburg, 16-th Line, 29),
performing final qualification work in the Group of Experimental Microbiology, D.O. Ott Reasearch Institute of Obstetrics, Gynecology and Reproductology
(199034, Russia, St. Petersburg, Mendeleyevskaya Line, 3), gimkolos@gmail.com
Kira V. Shalepo, PhD (Bio), Senior Researcher, Department of Medical Microbiology, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology,
199034, St. Petersburg, Mendeleevskaya line, 3; Associate Professor, Department of Clinical Laboratory Diagnostics, St. Petersburg State Pediatric Medical University,
Ministry of Health of Russia, 194100, Russia, St. Petersburg, Litovskaya str., 2, 2474151@mail.ru
Olga V. Budilovskaya, Researcher, Department of Medical Microbiology, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology,
199034, St. Petersburg, Mendeleevskaya line, 3; Assistant, Department of Clinical Laboratory Diagnostics, St. Petersburg State Pediatric Medical University,
Ministry of Health of Russia, 194100, Russia, St. Petersburg, Litovskaya str., 2, o.budilovskaya@gmail.com
Tatiana A. Khusnutdinova, Researcher, Department of Medical Microbiology, D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology,
199034, Russia, St. Petersburg, Mendeleevskaya line, 3; Assistant, Department of Clinical Laboratory Diagnostics, St. Petersburg State Pediatric Medical University,
Ministry of Health of Russia, 194100, Russia, St. Petersburg, Litovskaya str., 2, husnutdinovat@yandex.ru
Alevtina M. Savicheva, Merited Scholar of the Russian Federation, Dr. Med. Sci., Professor, Head of the Department of Medical Microbiology, D.O. Ott Research Institute
of Obstetrics, Gynecology and Reproductology, 199034, Russia, St. Petersburg, Mendeleevskaya line, 3; Head of the Department of Clinical Laboratory Diagnostics,
St. Petersburg State Pediatric Medical University, Ministry of Health of Russia, 194100, Russia, St. Petersburg, Litovskaya str., 2, savitcheva@mail.ru
Igor Yu. Kogan, Corresponding Member of the RAS, Dr. Med. Sci., Professor, Director, D.O. Ott Reasearch Institute of Obstetrics, Gynecology and Reproductology, +7(812)328-98-22, ovr@ott.ru, 199034, Russia, St. Petersburg, Mendeleyevskaya Line, 3.
Corresponding author: Elena V. Shipitsyna, shipitsyna@inbox.ru
Authors’ contributions: Shipitsyna E.V. – conception and design of the study, laboratory analysis, analysis of results, manuscript editing, approval of the final version of the article; Korkina S.A., Krysanova A.A., Kolousova K.A., Shalepo K.V.,
Budilovskaya O.V., Khusnutdinova T.A. – culturing bacterial strains, laboratory analysis, manuscript editing, approval of the final version of the article; Savicheva A.M. – analysis of the results, manuscript editing, approval of the final version of the article;
Kogan I.Yu. – analysis of the results, final manuscript editing and approval of the final version of the article.
Conflicts of interest: The authors have no conflicts of interest to declare.
Funding: The study was conducted within the framework of the state order of the Ministry of Science and Higher Education of the Russian Federation.
Authors’ Data Sharing Statement: The data supporting the findings of this study are available on request from the corresponding author after approval from the principal investigator.
For citation: Shipitsyna E.V., Korkina S.A., Krysanova A.A., Kolousova K.A., Shalepo K.V., Budilovskaya O.V., Khusnutdinova T.A., Savicheva A.M., Kogan I.Yu.
Sialidase activity of vaginal bacteria in reproductive-age women.
Akusherstvo i Ginekologiya/Obstetrics and Gynecology. 2022; 11: 122-130 (in Russian)
https://dx.doi.org/10.18565/aig.2022.11.122-130



