В настоящее время значительное число браков является бесплодными и их количество с каждым годом растет. При лечении различных видов бесплодия широкое распространение получили вспомогательные репродуктивные технологии (ВРТ), так как по сравнению с другими доступными методами лечения бесплодия ВРТ наиболее эффективны и имеют высокие показатели наступления беременности и рождения живых детей. Тем не менее, лишь 32,3% циклов ВРТ, проведенных впервые у женщин до 37 лет, заканчиваются рождением здорового ребенка. Частота рождения ребенка у пациенток старшего репродуктивного возраста в первом цикле ЭКО не превышает 12,3% [1].
Помимо правильно подобранной схемы стимуляции, положительный результат в программах ВРТ зависит от качества переносимого эмбриона и рецептивности эндометрия [2]. Имплантация эмбриона в эндометрий – многоэтапный процесс, опосредованный многоуровневой регуляцией внутри- и межклеточных взаимодействий, необходимой для дальнейшего развития бластоцисты, распознавания беременности и адаптации к ней организма матери.
Несмотря на то, что наступление беременности в программах ВРТ зависит от многих факторов, выбор наиболее качественного и жизнеспособного эмбриона для переноса в полость матки – одна из наиболее важных задач. Селекция эмбрионов с наибольшим имплантационным потенциалом, как правило, производится на основании визуальной оценки их морфологических свойств. Оценка морфологических параметров подразумевает измерение размера эмбриона, изучение его внутренней клеточной массы и трофэктодермы. Однако не все эмбрионы «хорошего» морфологического качества успешно имплантируются, в связи с чем возникает необходимость внедрения дополнительных неинвазивных технологий селективного выбора эмбриона с высоким имплантационным потенциалом [3]. Безусловно, нарушения имплантации могут быть связаны не только с «эмбриональными», но и с «материнскими» факторами. Материнские факторы включают анатомические аномалии матки (3,5% женщин, страдающих бесплодием), тромбофилию (20% женщин, страдающих бесплодием), нарушение рецептивности эндометрия (10–15% случаев бесплодия) и иммунологические факторы (5–15% случаев бесплодия в паре) [4].
За последние годы было изучено значительное число потенциальных биомаркеров качества эмбрионов и их имплантационной способности. Достаточно многообещающим методом выступает предимплантационный генетический скрининг (ПГС), но он остается дорогим и инвазивным. Поэтому продолжается поиск маркеров, которые дополнят общепринятый стандарт оценки качества эмбриона и его имплантационного потенциала. Основными требованиями, предъявляемыми к потенциальным маркерам, являются возможность оценки качества эмбриона без инвазивных вмешательств, четкие и воспроизводимые количественные характеристики, применимость в рутинной клинической практике [5].
Способы оценки качества эмбрионов в рутинной клинической практике
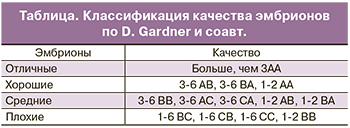 Жизнеспособность эмбрионов при культивировании зависит от трех ключевых параметров: температуры, рН и осмолярности культуральной среды. Развитие до стадии бластоцисты не обязательно происходит по строго определенному графику. Было показано, что скорость развития эмбриона коррелирует с его способностью к свободному выходу из прозрачной оболочки. Независимо от особенностей среды, в которой протекало развитие эмбриона, в идеале через 120 ч (на 5-й день) здоровый человеческий эмбрион должен достичь стадии бластоцисты. Развитие эмбриона на ранних стадиях обеспечивается благодаря трансляции материнских мРНК, происходящих из яйцеклетки. Собственный геном эмбриона человека активируется через несколько дней после оплодотворения. На 5 день своего развития эмбрион должен состоять из 50–200 клеток, 20–30% которых приходится на внутреннюю клеточную массу, а 70% – на трофобласт. В соответствии с рекомендациями Стамбульского Консенсуса, качественная оценка эмбрионов должна включать в себя морфокинетические характеристики и плоидность. В рутинной клинической практике для оценки качества эмбриона применяются морфологические критерии Gardner и соавт. (таблица) [6]. Данные критерии включают в себя оценку внутренней клеточной массы, трофэктодермального слоя, прозрачной оболочки, а также скорость развития бластоцисты.
Жизнеспособность эмбрионов при культивировании зависит от трех ключевых параметров: температуры, рН и осмолярности культуральной среды. Развитие до стадии бластоцисты не обязательно происходит по строго определенному графику. Было показано, что скорость развития эмбриона коррелирует с его способностью к свободному выходу из прозрачной оболочки. Независимо от особенностей среды, в которой протекало развитие эмбриона, в идеале через 120 ч (на 5-й день) здоровый человеческий эмбрион должен достичь стадии бластоцисты. Развитие эмбриона на ранних стадиях обеспечивается благодаря трансляции материнских мРНК, происходящих из яйцеклетки. Собственный геном эмбриона человека активируется через несколько дней после оплодотворения. На 5 день своего развития эмбрион должен состоять из 50–200 клеток, 20–30% которых приходится на внутреннюю клеточную массу, а 70% – на трофобласт. В соответствии с рекомендациями Стамбульского Консенсуса, качественная оценка эмбрионов должна включать в себя морфокинетические характеристики и плоидность. В рутинной клинической практике для оценки качества эмбриона применяются морфологические критерии Gardner и соавт. (таблица) [6]. Данные критерии включают в себя оценку внутренней клеточной массы, трофэктодермального слоя, прозрачной оболочки, а также скорость развития бластоцисты.
Согласно классификации Gardner и соавт. цифрой обозначена степень зрелости бластоцисты:
- 1-я степень – ранняя бластоциста, полость бластоцисты меньше половины объема эмбриона;
- 2-я степень – полость бластоцисты больше половины объема эмбриона;
- 3-я степень – полная бластоциста, полость полностью занимает объем эмбриона;
- 4-я степень – расширенная бластоциста, полость бластоцисты становится больше и начинает истончаться прозрачная оболочка;
- 5-я степень – трофэктодерма начинает проникать через прозрачную оболочку;
- 6-я степень — вылупившаяся бластоциста, покинувшая zona pellucida (ZP).
Первая буква в классификации характеризует состояние внутренней клеточной массы:
- А – большое количество плотно упакованных клеток;
- В – среднее количество клеток с более свободной упаковкой;
- С – незначительное количество клеток.
Вторая буква в классификации характеризует состояние трофэктодермального слоя:
- А – много клеток, формирующих трофэктодерму;
- В – немного клеток;
- С – незначительное количество больших клеток.
Стоит отметить, что неивазивных способов оценки качества эмбрионов и их жизнеспособности крайне мало. Возможный вариант – ПГС, но он остается трудоемким и дорогостоящим методом. Более того, 50% эмбрионов, которые по результатам ПГС оказываются эуплоидными, при переносе в полость матки не приводят к беременности [7]. Таким образом, на сегодняшний день в рутинной клинической практике для оценки жизнеспособности эмбриона используются морфокинетические критерии Gardner и соавт. и ПГС.
Новые подходы для определения фертильности на стадиях оогенеза, сперматогенеза и раннего эмбрионального развития с помощью «омиксных» технологий
Одним из перспективных подходов для неивазивной оценки качества эмбриона и его имплантационного потенциала является масс-спектрометрический (МС) анализ культуральной среды. МС метод позволяет оценить качество эмбриона по метаболомному и протеомному профилю среды его культивирования. МС – один из наиболее информативных методов для молекулярного профилирования путем измерения отношения массы к заряду ионов, образующихся из молекул анализируемой смеси; также при использовании меченых стандартов возможно и количественное определение молекулярных компонент в анализируемой пробе.
Анализ метаболомного профиля эмбриона
Метаболомика – это наука, изучающая функционирование живой системы путем анализа качественного и количественного состава метаболитов, являющихся субстратами, интермедиатами или продуктами большинства биохимических реакций. К метаболитам относятся нуклеотиды, составляющие основу генома и транскриптома; аминокислоты, необходимые для синтеза белков; липиды: холестерин, фосфолипиды, триглицериды, липопротеины, и др. [8]. Любые изменения метаболомного профиля отражают изменения функциональной активности живого объекта. Метаболомика может предоставить информацию для оценки функционального статуса половых клеток, эмбрионов и эндометрия, что позволит внести некие коррективы в лечение бесплодия супружеских пар, хотя в настоящее время остается много открытых и не решенных вопросов в этой области [9]. Из всего многообразия метаболитов, известных в настоящее время, наибольшее внимание уделяли углеводам и аминокислотам. Более 50 лет изучают возможность их использования для оценки физиологического состояния эмбрионов и прогнозирования вероятности их успешной имплантации.
На начальных стадиях эмбриогенеза делящиеся клетки в основном используют пируват, лактат и аминокислоты, в то время как на более поздних этапах развития эмбрионы начинают активно поглощать глюкозу [10]. Это наблюдение применили для оценки жизнеспособности коровьих бластоцист. Оказалось, что эмбрионы, у которых потребление глюкозы было выше 5 мкг/ч, обладали большим потенциалом развития, нежели эмбрионы с более низкими показателями поглощения глюкозы [11]. Аналогичным образом повышенная скорость поглощения глюкозы также была ассоциирована с более высокой эффективностью имплантации эмбрионов мыши и человека [12]. В то же время, в ходе дальнейших исследований авторы пришли к выводу, что запредельные показатели поглощения глюкозы наблюдаются у эмбрионов, потерявших способность регулировать свой метаболизм, что приводило к значительному ухудшению жизнеспособности. Напротив, эмбрионы, сохранившие способность контролировать метаболические процессы при наличии высокого уровня поглощения глюкозы, обладают высокой жизнеспособностью. В настоящее время анализ углеводов нельзя предложить для надежной оценки качества эмбрионов, так как пока не определены четкие количественные диапазоны потребления глюкозы эмбрионом, ассоциированные с высокой и низкой его жизнеспособностью.
Аминокислоты играют несколько ключевых ролей во время эмбриогенеза, в числе которых – обеспечение энергии, биосинтетических предшественников, буферов внутриклеточного рН, антиоксидантов и регуляторов дифференцировки [13–15]. Было показано, что добавление заменимых и незаменимых аминокислот в культуральную среду значительно увеличивает скорость перехода от ранней стадии дробления к стадии бластоцисты [16, 17]. Учитывая участие аминокислот в широком спектре важных метаболических функций, разумно предположить, что эмбрионы с наибольшим потенциалом развития будут иметь характерный профиль аминокислот. Первоначальные исследования, проведенные Хаутоном и его коллегами, в которых концентрации аминокислот измеряли в содержащей эмбрион культуральной среде, выявили более низкое поглощение глутамина, аргинина и метионина, а также более низкое выделение аланина и аспарагина у 2–3-дневных эмбрионов, способных в дальнейшем дорасти до стадии бластоцисты, по сравнению с неразвивающимися эмбрионами [18]. К сожалению, как и в случае анализа углеводного профиля, не удалось вынести окончательную оценку применимости уровня поглощения/выделения аминокислот эмбрионами с наибольшим потенциалом развития. Измерение уровня аминокислот, как и уровня углеводов в культуральной среде может, в конечном итоге, найти клиническое применение, но в настоящее время ограничивается только исследовательскими задачами. Два основных затруднения, которые стоят на пути внедрения анализа метаболитов в рутинную клиническую практику: 1) высокая зависимость скорости изменения метаболизма эмбрионов от состава культуральной среды и концентрации кислорода; 2) необходимость использования аналитических инструментов с высокой точностью и чувствительностью для определения незначительных изменений концентрации метаболитов в результате жизнедеятельности единичного эмбриона. Расхождения в получаемых данных разными научными коллективами в связи с использованием культуральных сред различного состава, а также разных МС- платформ не позволяют пока создать единый протокол по отбору эмбрионов с высокими показателями жизнеспособности по оценке уровня метаболитов в культуральной среде.
Анализ протеомного профиля эмбриона
МС анализ протеомного профиля эмбриона позволяет выявить новые диагностически значимые маркерные белки для выбора новых мишеней в лечении бесплодия.
На данный момент основные исследования по изучению протеомного состава бластоцисты проведены с использованием лабораторных животных, в частности, мышиных эмбрионов [19]. Однако существует ряд работ по исследованию единичных бластоцист человека [20]. Авторам данных работ удалось идентифицировать 182 белка бластоцели, которые можно разделить на три группы: 1) белки материнского происхождения OOEP, NLRP5, TLE6, PADI6, ECAT1, экспрессия которых важна при материнско-зиготическом переходе и сохраняется после активации эмбрионального генома; 2) белки, участвующие в локальном иммунном ответе и межклеточных взаимодействиях, например, S100A8 и S100A9, обладающие провоспалительной функцией и имеющие повышенный уровень при раннем прерывании беременности; 3) белки цитоплазматической локализации, участвующие в метаболических процессах. Кроме того, авторами этих работ был проведен корреляционный анализ уровня экспрессии мажорных белков (GAPDH, ACTA, H2A) бластоцели 13 эмбрионов с данными цитогенетического анализа, полученными в ходе бластоцентеза. Выявлены достоверные отличия в уровне экспрессии GAPDH (р=0,029) между эуплоидными и анеуплоидными эмбрионами. Несмотря на полученные оптимистичные результаты, авторами исследования подчеркивается необходимость подтверждения этих данных на независимой более многочисленной выборке.
В исследовании других авторов было проведено протеомное профилирование бластоцист человека для выявления возможных механизмов влияния возрастных параметров женщин репродуктивного возраста на их фертильность, а также для поиска потенциальных биомаркеров качества бластоцист и положительного исхода в программах ВРТ [21]. В данном эксперименте были проанализированы эмбрионы женщин разных возрастных групп (<37 лет, >37 лет). В обеих группах было идентифицировано 148 белков, среди которых 108 белков были описаны впервые в жидкости бластоцели. По уровню экспрессии 109 белков были выявлены отличия двух групп. Большинство дифференциально экспрессированных белков участвуют в процессах, связанных с убиквитинированием, среди которых убиквитин-конъюгирующий фермент (UBC), С-концевая убиквитин-гидролаза (UCHL1), RNF223, KLHL17, PSMD4 (компонент протеасомного комплекса 26S, отвечающий за распад убиквитинированных белков) и ZFAND6 (полиубиквитин-связывающий белок, участвующий в регуляции транскрипционного фактора NF-kB) имеют резко сниженный уровень экспрессии или она вовсе отсутствует, как в случае ZFAND6, в жидкости бластоцели женщин из второй возрастной группы (>37 лет). Авторы работы пришли к выводу, что дисрегуляция системы убиквитинирования белков в растущем эмбрионе может быть причиной неудач в программах ВРТ.
Было проведено исследование секретома эмбриона на ранних стадиях его развития [22]. Исследование было направлено на выявление потенциальных маркеров, характеризующих способность бластоцисты к эффективной имплантации. В эксперименте при использовании технологии микрочипов проводили сравнение протеомного профиля среды культивирования эмбрионов, обладающих доказанной впоследствии способностью имплантироваться в децидуальный слой матки и не обладавших такой активностью, относительно культуральной среды, не содержащей эмбрион. Было выявлено снижение уровней секреции GM-CSF и CXCL13 у эмбрионов, обладающих имплантационным потенциалом [23].
Изучение протеомного состава культуральной среды эмбрионов сопряжено с рядом технических трудностей, связанных с искусственным обогащением среды мажорными белками сыворотки для нормального развития эмбриона, что требует дополнительных этапов пробоподготовки, в ходе которых возможны потери потенциально значимых низкопредставленных белков. Среди потенциальных биомаркеров имплантационной способности эмбриона следует выделить такие белки, как интегрин альфа-X и FRAS1 [24]. Многообещающие результаты недавних исследований показали, что интерлейкин–6, обнаруженный в содержащей эмбрион культуральной среде, может выступать еще одним маркером имплантационного потенциала эмбриона. Было доказано, что уровень интерлейкина-6 значительно повышен в культуральной среде эмбрионов с подтвержденной имплантацией в децидуальный слой матки [25].
Анализ малых некодирующих РНК как основных регуляторов репродуктивной функции
Регуляция экспрессии генов на посттранскрипционном уровне является одним из механизмов, определяющих различный фенотип и функции клеток организма. Клетки и ткани, отвечающие за выполнение репродуктивных функций, уникальны в том, что они непрерывно подвергаются существенной реорганизации, как на транскриптомном, так и на протеомном уровнях во время гаметогенеза и эмбриогенеза. Временные изменения протеомного профиля под контролем не кодирующих белок РНК в последние десятилетия находятся в поле пристального внимания ученых-репродуктологов. Принято считать, что некодирующие РНК, такие как транспортные РНК, рибосомальные РНК, малые ядерные и малые ядрышковые РНК, не являются регуляторными и главным образом поддерживают функцию клеток без весомого вклада в определение ее фенотипа. Другие классы некодирующих РНК, а именно: микроРНК (miRNAs), эндогенные малые интерферирующие РНК (endo-siRNAs) и PIWI-взаимодействующие РНК (piRNAs) выполняют регуляторную роль, что в значительной степени влияет на фенотип и функцию клеток. Основными методами анализа уровня экспрессии РНК являются технологии, основанные на гибридизации флуоресцентно-меченого зонда с РНК-мишенью, метод глубокого секвенирования и количественная полимеразная цепная реакция в реальном времени, сопряженная с обратной транскрипцией.
Биогенез и механизм действия малых регуляторных некодирующих РНК микроРНК (miRNAs)
MiRNAs – эндогенные, короткие, не кодирующие белок молекулы РНК длиной 21–24 нуклеотида, присущие всем многоклеточным организмам и регулирующие экспрессию гена на пост-транскрипционном уровне путем связывания с комплементарными последовательностями мРНК-мишени [26]. Большинство miRNAs транскрибируется РНК-полимеразой II с образованием первичного транскрипта (pri-miRNAs) длиной 2-4 тысячи оснований с 5’-7-метил-гуанозиновой кэп-структурой, 3’-поли(А)-хвостом и сложной вторичной структурой. Дальнейший процессинг pri-miRNAs происходит в ядре под действием высоко консервативной РНКазы III Drosha в комплексе с кофактором DGCR8 с формированием 70–100 нуклеотидной шпилечной структуры – предшественника miRNA (pre-miRNA). Последний активно экспортируется в цитоплазму с участием экспортина-5 в комплексе с RAN-ГТФазой, где подвергается процессингу с образованием двухцепочечной зрелой miRNA в среднем длиной 22 нуклеотида под действием РНКазы III Dicer в комплексе с Tar РНК-связывающим белком (TRBP). Впоследствии, одна цепь дуплекса miRNA встраивается в мультибелковый (белки Argonaute) РНК-ингибирующий комплекс (RISC) для целенаправленного транспорта RISC к мРНК-мишени и блокирования ее трансляции, в то время как другая цепь (miRNA*) подвергается деградации [27].
Эндогенные малые интерферирующие РНК (endo-siRNAs)
siRNAs образуются из эндогенных двухцепочечных РНК-предшественников в результате двунаправленной транскрипции генов/псевдогенов или транскрипции инвертированных повторов с последующим процессингом рибонуклеазой III Dicer без предшествующего процессинга с участием Drosha [28], что отличает биогенез siRNAs от такового miRNAs. Процессированные молекулы siRNAs образуют комплекс с AGO-белками, обеспечивающими взаимодействие направляющей цепи siRNA c мРНК-мишенью с последующим эндонуклеазным расщеплением и деградацией мРНК.
PIWI-взаимодействующие РНК (piRNAs)
Биогенез piRNAs менее изучен и понятен, нежели таковой miRNAs и siRNAs. PIWI-взаимодействующие РНК являются продуктом расщепления длинных, одноцепочечных РНК-предшественников посредством Dicer-независимого механизма [29]. По данным глубокого секвенирования, существует несколько разновидностей piRNAs [30]: одна фракция piRNAs происходит из кластеризованных геномных локусов в повторяющихся последовательностях (Класс I), вторая фракция piRNAs является продуктом расщепления РНК транспозонов (Класс II). Оставшаяся фракция piRNAs происходит из различных геномных областей, в том числе 3’-нетранслируемой области некоторых мРНК, предполагая наличие у piRNAs функции регулятора уровня экспрессии гена, помимо контроля хромосомных перестроек путем репрессии транспозонов. Пополнение пула piRNAs и одновременная репрессия транспозонов происходит по механизму «пинг-понг» [31]. Белки PIWI и AUB связывают антисмысловую первичную piRNA, синтезированную на кластеризованных геномных локусах, с формированием комплексов Piwi-piRISCs и Aub-piRISCs, соответственно. Piwi-piRISCs импортируется в ядро и блокирует экспрессию транспозона путем тройного метилирования лизина 9 гистона Н3 (Н3К9me3) и локального формирования гетерохроматина. Aub-piRISCs связывается с мРНК транспозона и расщепляет ее с образованием смысловой вторичной piRNA. Последняя образует комплекс с AGO3 для связывания с антисмысловой первичной piRNA, синтезированной с геномного кластера, для пополнения пула первичных piRNAs.
Участие малых некодирующих РНК в гаметогенезе
Оогенез
Оогенез млекопитающих включает в себя 3 фазы – митотическое деление овогоний (эмбриональный период до 1 года жизни), фаза роста (образование ооцитов 1-го порядка), фаза созревания (два меойтических деления). Доказано, что сразу после оплодотворения ооцит транскрипционно неактивен; активация эмбрионального генома происходит только на стадии двух-восьми-клеточного уровня развития в зависимости от вида млекопитающих. Отсутствие способности транскрибировать мРНК на ранних стадиях развития навело на мысль различных ученых о возможной посттранскрипционной регуляции экспрессии генов с появлением работ по анализу пула некодирующих РНК в созревающем ооците. Была обнаружена экспрессия miRNAs, endo-siRNAs и piRNAs в ооцитах различных видов млекопитающих на разных стадиях развития [32]. В результате секвенирования малых некодирующих РНК в ооцитах и клетках кумулюса свиньи во время фазы созревания in vitro было выявлено значительное повышение экспрессии miR-21 и снижение экспрессии miR-574-3p [33].
Ингибирование экспрессии miR-21 негативно влияет на мейотическое деление и достижение метафазы-II, подавляет овуляцию и индуцирует апоптоз, что возможно объясняется повышением уровня экспрессии гена-мишени miR-21 – PDCD4 (Programmed cell death-4) – регулятора апоптотического процесса. Что касается piRNA, наиболее представленными оказались 63 транскрипта, кодируемые 6 хромосомой (участок 246415-259463), среди которых наибольший уровень экспрессии был выявлен у piR84651 (41,6%-80,7% от общего числа прочтений piRNA в зависимости от стадии созревания ооцита и эмбриона).
Важно отметить, что у разных видов млекопитающих вклад малых некодирующих РНК в оогенез и раннее эмбриональное развитие различен. Хотя miRNAs в большом изобилии экспрессируются в ооцитах мышей, было доказано, что Dgcr8 не влияет ни на регуляцию уровня экспрессии мРНК, ни на созревание ооцитов, в то время как Dicer и Ago2 необходимы для полноценного оогенеза [26, 34]. Эти исследования позволили предположить, что ключевую роль в развитии и созревании ооцитов у мышей играют endo-siRNA или синтезируемые неканоническим путем (Dgcr8-независимым) miRNAs [35]. Кроме того, в ооцитах мышей выявлена экспрессия белков Piwi, но ни один из них не оказывает влияния на фертильность самок, так же, как и piRNAs, уровень экспрессии которых ничтожно мал. Напротив, Elke F. Roovers с коллегами представил доказательства выраженной экспрессии PIWIL1, PIWIL2, PIWIL3 и piRNAs в ооцитах человека, быка и макаки, причем спектр piRNAs в ооцитах сходен с таковым в сперматозоидах яичек на пахитенной стадии мейоза, и их основная роль заключается в регуляции экспрессии транспозонов [36]. В таких модельных организмах, как Drosophila и Zebrafish, белки PIWI и piRNAs существенно важны, как во время оогенеза, так и сперматогенеза [37].
Сперматогенез
Сперматогенез является высоко регулируемым процессом, в основном, на транскрипционном и пост-транскрипционном уровнях, важную роль в котором играют малые некодирующие РНК. Сперматогенез состоит из трех фаз: 1) самообновление сперматогоний путем митотических делений, 2) дифференцировка сперматогоний с образованием первичных сперматоцитов, которые проходят два мейотических деления для образования вторичных сперматоцитов и гаплоидных сперматид, 3) морфологические изменения гаплоидных сперматид с образованием зрелого сперматозоида. Недавние исследования выявили регуляторную роль определенного спектра miRNAs на разных стадиях сперматогенеза. Так, например, в поддержании популяции стволовых клеток сперматогоний существенную роль играют miR-21, Mir-17-92 (Mirc1) и Mir-106b-25 (Mirc3) кластеров, Mir146, miR-221/222, miR-135a и ее ген-мишень FOXO1 [38]. Важную роль в инициации дифференцировки сперматогоний играет семейство Mirlet7 [39]. Для сперматогоний и первичных сперматоцитов характерна экспрессия miR-383, влияющая на фертильность мужских особей. Другие miRNAs специфичным образом функциональны во время профазы мейоза, а именно: miR-214, miR-24, miR-206, miR-202, miR-298 [40]. Важную роль в инициации/предотвращении апоптоза во время мейотической гомологичной рекомбинации сперматоцитов играют кластер miR-449, кластер miR-17-92 и miR-34b/c, воздействуя либо на транскрипционный фактор E2F-1, либо на фактор активации транскрипции 1 ATF1 [41]. Некоторые виды miRNAs участвуют в регуляции экспрессии генов в постмейотических сперматидах. Например, miR-122a и miR-469 обусловливают снижение уровня замещающего белка Tnp2, ядерного белка, вовлеченного в перестройку хроматина при замещении гистаминов на протамины для обеспечения возможности морфологических изменений ядра во время спермиогенеза [42, 43]. Дисрегуляция удлинения сперматидов за счет структурных цитоплазматических и ядерных перестроек приводит к азооспермии.
В отличие от miRNAs, которая высоко представлена на всех стадиях сперматогенеза, пик экспрессии piRNAs приходится только на стадию образования пахитенных сперматоцитов и круглых сперматидов. Пахитенные piRNA отличаются от других видов piRNAs тем, что образуются по основному пути биогенеза независимо от «пинг-понг»-механизма амплификации piRNAs (см. выше) [44]. Функциональное значение такого всплеска экспрессии пахитенных piRNAs интенсивно анализируется. Известно, что piRNAs участвуют в MIWI-опосредованной регуляции экспрессии транспозонов. Однако популяция пахитенных piRNAs гетерогенна и только 20% piRNAs картируется на гены транспозонов. Большинство пахитенных piRNAs кодируется межгенными участками, не содержащими повторов, а также кодирующими белок генами, причем для этой популяции piRNAs не было найдено РНК-мишеней [45]. A. Vourekas и соавт. предположили, что эта фракция piRNAs является продуктом деградации мейотических РНК, не требующихся более в гаплоидных клетках. Роль piRNAs и ассоциированных с ними белков PIWI на других стадиях сперматогенеза подробным образом описана в обзорной статье [46], в которой подчеркивается роль piRNAs в эпигенетической модификации ДНК, регуляции трансляции мРНК и ее стабильности, поддержании и самообновлении герминальных стволовых клеток.
Роль малых некодирующих РНК в эмбриональном развитии
Успешное эмбриональное развитие сразу после оплодотворения зависит от координированной реализации программ по уничтожению материнских мРНК и активации зиготического генома с последующим синтезом эмбриональных мРНК и трансляцией на них белков. Было обнаружено, что piRNA играют важную роль в материнско-зиготическом переходе [47]. Для уничтожения материнской мРНК активируется процесс ее деаденилирования, участником которого являются piRNA путем взаимодействия с основными белками ферментативного комплекса – деаденилазой CCR4 и РНК-связывающим белком Smaug [48]. Было обнаружено, что ингибирование piRNA приводит к нарушению формирования эмбриона и вызывает выраженные дефекты головной части плода [49]. В некоторых организмах роль регулятора материнско-зиготического перехода отводится также miRNAs и siRNAs [50, 51]. У разных животных материнско-зиготический переход происходит в разное время. Например, у млекопитающих активация эмбриональных генов начинается после первого деления зиготы, а у рыб только после 10 делений [52].
Результатом недавних исследований явилось доказательство использования специфических miRNА и piRNA в качестве маркеров хромосомного набора эмбриона и его имплантационного потенциала [53]. Rosenbluth и соавт. обнаружили, что профиль экспрессии miRNА различается в эуплоидных и анеуплоидных эмбриональных клетках, а именно: в анэуплоидных эмбрионах экспрессируется повышенное количество miR-372 и miR-191 [54].
Обнаружение специфических miRNА не только в эмбриональных клетках, но и в культуральной среде эмбрионов на стадии бластоцисты позволило рассматривать miRNА в качестве удобного неинвазивного маркера плоидности генома эмбриона [55].
Авторы данного исследования отмечают, что уровень экспрессии miR-20a и miR-30c напрямую коррелирует с частотой имплантации эмбриона. В свою очередь, Borges Jr и соавт. обнаружили, что повышенное содержание miR-142-3p в культуральной среде эмбриона свидетельствует о предстоящем нарушении процесса имплантации бластоцисты при ее переносе в полость матки [56].
В зависимости от стадии развития эмбриона концентрация miRNА в культуральной среде варьирует [57].
Спектр miRNА, полученный из культуральной среды морулы и эмбрионов более ранней стадии развития, был практически идентичен таковому в культуральной среде без эмбрионов. При этом 4-клеточный эмбрион в 2 раза более активно секретирует miRNА, нежели 2-клеточный эмбрион. По мере развития эмбриона секреция miRNА в культуральную среду усиливается. Таким образом, наиболее специфичными выступают miRNА, полученные из культуральной среды эмбрионов на более поздних стадиях развития.
Заключение
Среда культивирования эмбрионов различных стадий развития равно как и жидкость бластоцели – уникальные объекты исследования, содержащие информацию об энергетической, метаболической активности и функциональном состоянии внутриклеточных сигнальных систем эмбриона.
Прорывные исследования в области анализа профилей белков, метаболитов и малых ncRNA, секретируемых эмбрионом, и их сопоставление с возрастными особенностями супружеской пары, цитогенетическими данными эмбриона и его имплантационной способностью являются отправной точкой для создания диагностических и прогностических тест-систем по оценке качества эмбрионов в рамках проведения программ ВРТ. На сегодняшний день актуальной остается задача создания молекулярного портрета эмбриона с различными морфометрическими показателями. С этой целью использование анализа профиля экспрессии малых ncRNA чрезвычайно перспективно для внедрения в рутинную клиническую практику, так как, с одной стороны, малые ncRNA являются одними из основных регуляторов клеточного метаболизма. С другой стороны, РНК – широко используемый объект медицинской диагностики и в настоящий момент доступен разнообразный спектр отработанных высокочувствительных и специфичных методик по анализу уровня экспрессии РНК, огромный ассортимент аппаратной и реагентной базы. Кроме того, РНК-диагностика служит объективным, автоматизируемым, алгоритмизуемым методом и сочетает в себе все основные качества, предъявляемые к наиболее перспективным способам диагностики. Прогнозирование результатов программ ВРТ по профилю экспрессии малых ncRNA в культуральной среде эмбрионов обеспечивает уникальную возможность неинвазивной и ранней диагностики имплантационного потенциала бластоцисты. Именно оптимизация выбора наилучших эмбрионов с максимальной способностью к имплантации обещает стать отправной точкой в повышении эффективности программ ВРТ.



